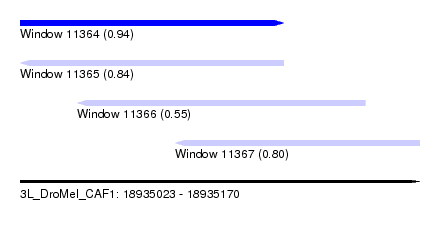
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 18,935,023 – 18,935,170 |
| Length | 147 |
| Max. P | 0.944343 |

| Location | 18,935,023 – 18,935,120 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 77.29 |
| Mean single sequence MFE | -17.14 |
| Consensus MFE | -11.79 |
| Energy contribution | -12.48 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.34 |
| SVM RNA-class probability | 0.944343 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
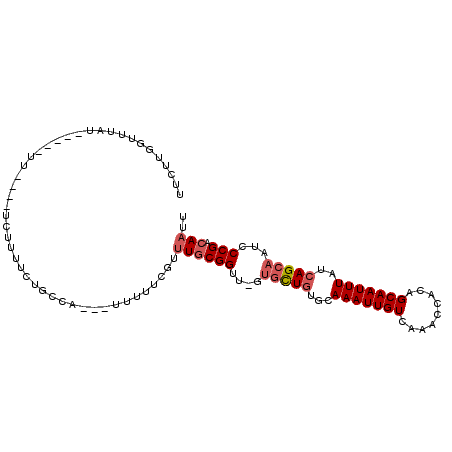
>3L_DroMel_CAF1 18935023 97 + 23771897 UUCUUGGUUUAU-----UUACACUCUUUUCUGCCA---UUUUUCGUUUUCGGUU-GUGCUGUGCAAAUUGUCAAACCACAGCAAUUUAUCAGCAAUCCCGACAAUU ....((((....-----..............))))---......(((.((((..-.(((((...(((((((.........)))))))..)))))...)))).))). ( -18.47) >DroPse_CAF1 76141 88 + 1 UUCUGUUUU---------------UUUUUUUACAG---UUUUUCUACUGCGGUUUUUGCUGUGCAAAUUGUCAAACCACAGCAAUUUAUCAGCAAUCCCGACAAUU ...(((...---------------........(((---(......))))(((...((((((...(((((((.........)))))))..))))))..))))))... ( -18.00) >DroSim_CAF1 70549 97 + 1 UUCUUGGUUUAU-----UUGCACUCCUUUCUGCCA---UUUUUCGUUUGCGGUU-GUGCUGUGCAAAUUGUCAAACCACAGCAAUUUAUCAGCAAUCCCGACAAUU ....((((....-----..............))))---........((((((..-.(((((...(((((((.........)))))))..)))))...))).))).. ( -18.47) >DroEre_CAF1 70848 102 + 1 UUCCUGUUUUAUAGUACUUCCACUCUUUUCUGCCA---UUUUUCGCUUGCGGUU-GUGCUGUGCAAAUUGUCAAACCACAGCAAUUUAUCAGCAAUCCCGACAAUU ...................................---........((((((..-.(((((...(((((((.........)))))))..)))))...))).))).. ( -16.50) >DroWil_CAF1 91981 87 + 1 AUUUUUGCUUAU-----UU---------UGUUGUA----UUUUUAUUUGCGGCU-UUGUUCUGCAAAUUGUCAAACCACAGCAAUUUAUCAGCAAUCCCGACAAUU ....(((((((.-----.(---------((((((.----.(((.((((((((..-.....))))))))....)))..)))))))..))..)))))........... ( -17.90) >DroAna_CAF1 68203 88 + 1 UUAU-------------UU----CCUUUUUUGCCAAUUUUUUUCCAUUGCGGUU-UGCCUGUGCAAAUUGUCAAACCACAGCAAUUUAUCAGCAAUCCCGACAAUU ....-------------..----......................(((((((.(-(((.((...(((((((.........)))))))..))))))..))).)))). ( -13.50) >consensus UUCUUGGUUUAU_____UU____UCUUUUCUGCCA___UUUUUCGUUUGCGGUU_GUGCUGUGCAAAUUGUCAAACCACAGCAAUUUAUCAGCAAUCCCGACAAUU ..............................................((((((....(((((...(((((((.........)))))))..)))))...))).))).. (-11.79 = -12.48 + 0.70)


| Location | 18,935,023 – 18,935,120 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 77.29 |
| Mean single sequence MFE | -18.64 |
| Consensus MFE | -11.92 |
| Energy contribution | -12.62 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.842259 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
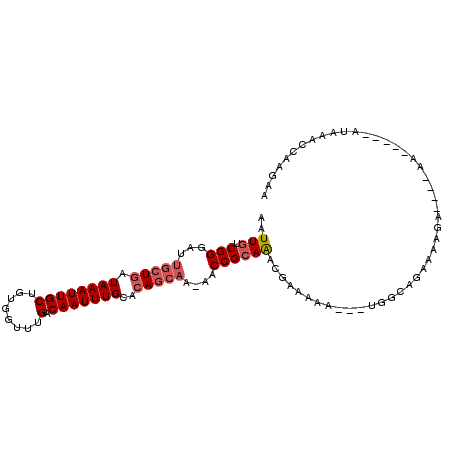
>3L_DroMel_CAF1 18935023 97 - 23771897 AAUUGUCGGGAUUGCUGAUAAAUUGCUGUGGUUUGACAAUUUGCACAGCAC-AACCGAAAACGAAAAA---UGGCAGAAAAGAGUGUAA-----AUAAACCAAGAA .....((((...(((((.((((((((........).)))))))..))))).-..))))..........---..................-----............ ( -16.70) >DroPse_CAF1 76141 88 - 1 AAUUGUCGGGAUUGCUGAUAAAUUGCUGUGGUUUGACAAUUUGCACAGCAAAAACCGCAGUAGAAAAA---CUGUAAAAAAA---------------AAAACAGAA ..(((((((.....))))))).((((((((((((......((((...)))))))))))))))).....---((((.......---------------...)))).. ( -23.00) >DroSim_CAF1 70549 97 - 1 AAUUGUCGGGAUUGCUGAUAAAUUGCUGUGGUUUGACAAUUUGCACAGCAC-AACCGCAAACGAAAAA---UGGCAGAAAGGAGUGCAA-----AUAAACCAAGAA ..(((((((.....))))))).......((((((....((((((((.((..-....))....(.....---...)........))))))-----)))))))).... ( -20.00) >DroEre_CAF1 70848 102 - 1 AAUUGUCGGGAUUGCUGAUAAAUUGCUGUGGUUUGACAAUUUGCACAGCAC-AACCGCAAGCGAAAAA---UGGCAGAAAAGAGUGGAAGUACUAUAAAACAGGAA ..((((((...(((((.......(((((((.............))))))).-.......)))))....---)))))).....((((....))))............ ( -19.38) >DroWil_CAF1 91981 87 - 1 AAUUGUCGGGAUUGCUGAUAAAUUGCUGUGGUUUGACAAUUUGCAGAACAA-AGCCGCAAAUAAAAA----UACAACA---------AA-----AUAAGCAAAAAU ..(((((((.....))))))).((((((((((((................)-))))))..((....)----)......---------..-----...))))).... ( -14.39) >DroAna_CAF1 68203 88 - 1 AAUUGUCGGGAUUGCUGAUAAAUUGCUGUGGUUUGACAAUUUGCACAGGCA-AACCGCAAUGGAAAAAAAUUGGCAAAAAAGG----AA-------------AUAA ..(((((((.....))))))).(((((((((((((.(..........).))-))))))(((........))))))))......----..-------------.... ( -18.40) >consensus AAUUGUCGGGAUUGCUGAUAAAUUGCUGUGGUUUGACAAUUUGCACAGCAA_AACCGCAAACGAAAAA___UGGCAGAAAAGA____AA_____AUAAACCAAGAA ..(((.(((...(((((.((((((((........).)))))))..)))))....)))))).............................................. (-11.92 = -12.62 + 0.69)

| Location | 18,935,044 – 18,935,150 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.06 |
| Mean single sequence MFE | -27.02 |
| Consensus MFE | -13.53 |
| Energy contribution | -13.98 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.554261 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
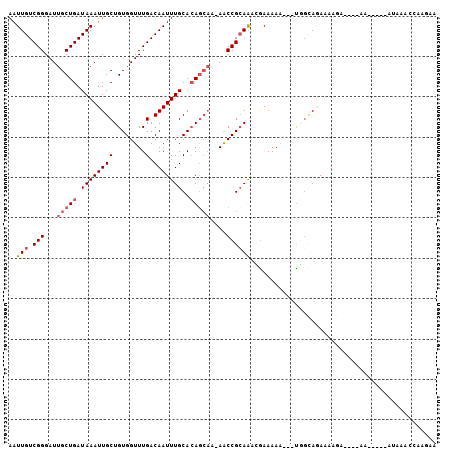
>3L_DroMel_CAF1 18935044 106 - 23771897 U----------GGCCUCUAAGCGCCUGCCAUUAAUAAUGCAAUUGUCGGGAUUGCUGAUAAAUUGCUGUGGUUUGACAAUUUGCACAGCAC-AACCGAAAACGAAAAA---UGGCAGAAA .----------.((......))..((((((((..........(((((((.....)))))))..(((((((.............))))))).-..............))---))))))... ( -27.42) >DroVir_CAF1 78299 97 - 1 ------------G--UCUAAG---CUGCCAUUAAUAAUGCAAUUGUCGGUGAAGCCGAUAAAUUGCUGUGGCUUGACAAUUUGCAGUG-CC-AACCGCAAUGCAAUAA----AACACACG ------------(--((.(((---(..(..........(((((((((((.....))))).)))))).)..)))))))...(((((.((-(.-....))).)))))...----........ ( -30.10) >DroGri_CAF1 67190 98 - 1 ------------G--UCUAAA---CUGCCAUUAAUAAUGCAAUUGUCGGUACAGCUGAUAAAUUGCAGUGGGUUGACAAUUUCCAGAGGCA-AACCGCAAUGAAAAUA----AACACACA ------------.--......---.((((........((((((((((((.....))))).))))))).((((.........))))..))))-................----........ ( -20.80) >DroWil_CAF1 91995 103 - 1 U----------GGUCUCUAAGCGCUUGCCAUUAAUAAUGCAAUUGUCGGGAUUGCUGAUAAAUUGCUGUGGUUUGACAAUUUGCAGAACAA-AGCCGCAAAUAAAAA----UACAACA-- (----------(((............))))........(((((((((((.....))))).))))))((((((((................)-)))))))........----.......-- ( -19.29) >DroAna_CAF1 68212 109 - 1 U----------GGUCUCUAAGCGCCUGCCAAUAAUAAUGCAAUUGUCGGGAUUGCUGAUAAAUUGCUGUGGUUUGACAAUUUGCACAGGCA-AACCGCAAUGGAAAAAAAUUGGCAAAAA .----------(((........)))(((((((......(((((((((((.....))))).))))))(((((((((.(..........).))-)))))))..........))))))).... ( -31.00) >DroPer_CAF1 77228 117 - 1 UGGUGUGCUGUGGUCUCUAAGCGCCUGCCAUUAAUAAUGCAAUUGUCGGGAUUGCUGAUAAAUUGCUGUGGUUUGACAAUUUGCACAGCAAAAACCGCAGUAGAAAAA---CUGUAAAAA .(((.(((((((((......))((..(((((.......(((((((((((.....))))).)))))).)))))..).)......)))))))...)))(((((......)---))))..... ( -33.50) >consensus U__________GGUCUCUAAGCGCCUGCCAUUAAUAAUGCAAUUGUCGGGAUUGCUGAUAAAUUGCUGUGGUUUGACAAUUUGCACAGCAA_AACCGCAAUGGAAAAA___UAGCACAAA ......................................(((((((((((.....))))).))))))(((((((...................)))))))..................... (-13.53 = -13.98 + 0.44)



| Location | 18,935,080 – 18,935,170 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 80.32 |
| Mean single sequence MFE | -26.02 |
| Consensus MFE | -17.19 |
| Energy contribution | -16.72 |
| Covariance contribution | -0.47 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.798972 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18935080 90 - 23771897 --------ACUGU--------UGUCC--ACAGCAUGUGUGGCCUCUAAGCGCCUGCCAUUAAUAAUGCAAUUGUCGGGAUUGCUGAUAAAUUGCUGUGGUUUGACAAU --------..(((--------((.((--((((((((.((((((.....).))).))))............(((((((.....)))))))..))))))))..))))).. ( -26.70) >DroVir_CAF1 78333 83 - 1 --------CCCGU--------UGUCC--ACUGCAUGUG--G--UCUAAG---CUGCCAUUAAUAAUGCAAUUGUCGGUGAAGCCGAUAAAUUGCUGUGGCUUGACAAU --------...((--------(((((--((.....)))--.--......---..(((((.......(((((((((((.....))))).)))))).)))))..)))))) ( -25.80) >DroGri_CAF1 67225 78 - 1 ---------------------UGUUG--ACUGCAUGUG--G--UCUAAA---CUGCCAUUAAUAAUGCAAUUGUCGGUACAGCUGAUAAAUUGCAGUGGGUUGACAAU ---------------------(((..--(((.((((((--(--(.....---..)))))......((((((((((((.....))))).)))))))))))))..))).. ( -24.30) >DroEre_CAF1 70910 98 - 1 --------ACUGUUGUCCUGUUGUCC--GCAGCAUGUGUGGCCUCUAAGCGCCUGCCAUUAAUAAUGCAAUUGUCGGGAUUGCUGAUAAAUUGCUGUGGUUUGACAAU --------...((((((.......((--((((((((.((((((.....).))).))))............(((((((.....)))))))..))))))))...)))))) ( -27.50) >DroWil_CAF1 92028 100 - 1 UUUGUCCUCCUCU--------UGUCCACACAGCAUGUGUGGUCUCUAAGCGCUUGCCAUUAAUAAUGCAAUUGUCGGGAUUGCUGAUAAAUUGCUGUGGUUUGACAAU .(((((.......--------.(.((((((.....)))))).)...........(((((.......(((((((((((.....))))).)))))).)))))..))))). ( -27.30) >DroAna_CAF1 68251 90 - 1 --------ACCGA--------UGUCC--ACAGCAUGUGUGGUCUCUAAGCGCCUGCCAAUAAUAAUGCAAUUGUCGGGAUUGCUGAUAAAUUGCUGUGGUUUGACAAU --------..(((--------...((--((((((......(((..(((...((((.((((.........)))).)))).)))..)))....)))))))).)))..... ( -24.50) >consensus ________ACUGU________UGUCC__ACAGCAUGUGUGG_CUCUAAGCGCCUGCCAUUAAUAAUGCAAUUGUCGGGAUUGCUGAUAAAUUGCUGUGGUUUGACAAU .....................((((...((((((...................(((..........))).(((((((.....)))))))..)))))).....)))).. (-17.19 = -16.72 + -0.47)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:35:24 2006