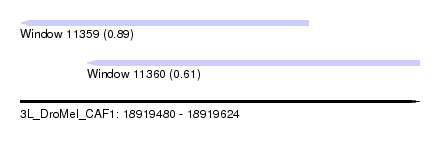
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 18,919,480 – 18,919,624 |
| Length | 144 |
| Max. P | 0.889875 |

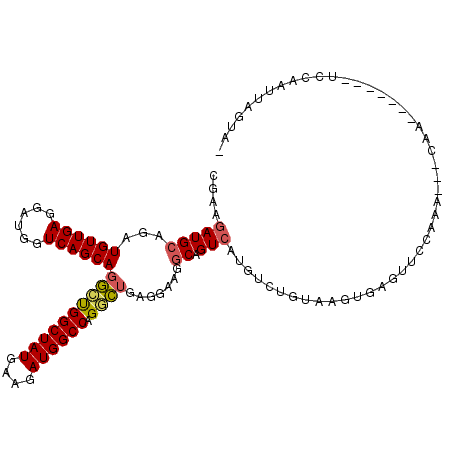
| Location | 18,919,480 – 18,919,584 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 77.93 |
| Mean single sequence MFE | -31.47 |
| Consensus MFE | -17.99 |
| Energy contribution | -18.02 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.889875 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18919480 104 - 23771897 CGAAGAUGCUGAUGUUGAGGAUGGUCAGCAGGCUGGCUAUGAAGAUGGCCAGGCUGAGGAAGGCGGUCAUGUCUGUAAGUGAGUUCCCAG---AAA-------UCCAAUUAGUU- .......((((((.....((((..((..(((.((((((((....)))))))).)))..)).((.(.((((........)))).).))...---..)-------))).)))))).- ( -31.70) >DroVir_CAF1 58145 113 - 1 GGUAUAUGGAAAUGUUGAGUAUGGUCAGCAAUGCGGCUAUGAAGAUGGCCAGCGAGAGAAAGGCCGUCAUGUCUGUGAGAGA--UAAAAAAAUAAACAGGUUAUUCAAUUAGCUU .((((.......((((((......))))))))))((((((((((((((((..(....)...))))))).(((((.....)))--)).................))))..))))). ( -28.71) >DroPse_CAF1 58506 102 - 1 AGUAGAUGCAGAUGUUGAGGAUGGUCAGCAGGCUGGCUAUGAAAAUGGCCAGGCUAAGGAAGGCAGUCAUAUCUGA--GGGAUGUGCACA---CAA-------CCGUAUUAGUA- ....(((((...((((((......))))))((((((((((....))))))))(((......))).((((((((...--..)))))).)).---...-------)))))))....- ( -33.10) >DroEre_CAF1 54985 104 - 1 CGAAGAUGCUGAUGUUGAGGAUGGUCAGCAGGCUGGCUAUGAAGAUGGCCAGGCUGAGGAAGGCCGUCAUGUCUGUAAGUGAGUUCCCGA---GAA-------UCCAAUUAGUA- ......(((((((......(((((((..(((.((((((((....)))))))).))).....)))))))...........((..(((....---)))-------..)))))))))- ( -34.70) >DroYak_CAF1 53501 104 - 1 CAAAGAUGCUGAUGUUGAGGAUGGUCAGCAGGGUGGCUAUGAAGAUGGCCAGGCUGAGGAAGGCAGUCAUGUCUGUAAGUGAGUUCCCAA---CAG-------UCCAAUAAGUA- ......((((..(((((.(((.(.((((((((.(((((((....)))))))(((((.......)))))...)))))...))).)))))))---)).-------.......))))- ( -28.90) >DroPer_CAF1 59361 102 - 1 AGUAGAUGCAGAUGUUGAGGAUGGUCAGCAGGCUGGCUAUGAAAAUGGCCAGGCUAAGGAACGCAGUCAUAUCUGA--GGGAUGUGCACA---CAA-------CCGUAUUAGUA- ....(((((...((((((......))))))((((((((((....))))))))((........)).((((((((...--..)))))).)).---...-------)))))))....- ( -31.70) >consensus CGAAGAUGCAGAUGUUGAGGAUGGUCAGCAGGCUGGCUAUGAAGAUGGCCAGGCUGAGGAAGGCAGUCAUGUCUGUAAGUGAGUUCCAAA___CAA_______UCCAAUUAGUA_ ....(((((...((((((......))))))((((((((((....)))))).)))).......)).)))............................................... (-17.99 = -18.02 + 0.03)

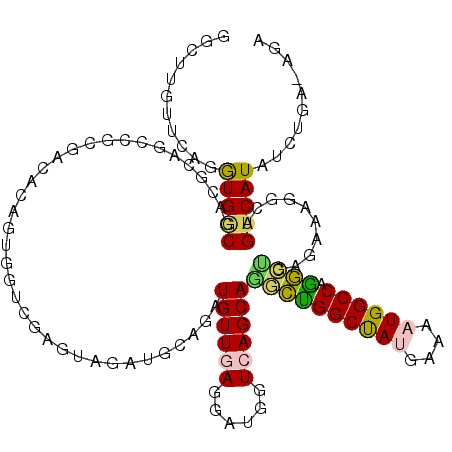
| Location | 18,919,504 – 18,919,624 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.06 |
| Mean single sequence MFE | -41.52 |
| Consensus MFE | -20.52 |
| Energy contribution | -19.63 |
| Covariance contribution | -0.89 |
| Combinations/Pair | 1.43 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.49 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.614951 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18919504 120 - 23771897 GGCUUGUCCAGGUGACGACGCAGCCGAGAGACUGUCGUUGCGAAGAUGCUGAUGUUGAGGAUGGUCAGCAGGCUGGCUAUGAAGAUGGCCAGGCUGAGGAAGGCGGUCAUGUCUGUAAGU ((((((((..(....))))).))))((.((((((((.((.(......((((((..........))))))((.((((((((....)))))))).))..).))))))))).).))....... ( -45.00) >DroVir_CAF1 58178 120 - 1 GGUUUGUUCAAAUGACGACGCAGCCGCGAUACGGUCGUCAGGUAUAUGGAAAUGUUGAGUAUGGUCAGCAAUGCGGCUAUGAAGAUGGCCAGCGAGAGAAAGGCCGUCAUGUCUGUGAGA ......((((...((((.((.(((((((((((.........)))).......((((((......)))))).))))))).))..(((((((..(....)...))))))).))))..)))). ( -39.70) >DroPse_CAF1 58530 118 - 1 GGCUUGUUCAGGUGGCGCCUCAGACGCGACACUGUGGUCGAGUAGAUGCAGAUGUUGAGGAUGGUCAGCAGGCUGGCUAUGAAAAUGGCCAGGCUAAGGAAGGCAGUCAUAUCUGA--GG ......((((((((((((((((..(.((((((((((.((.....))))))).))))).)..)).((...((.((((((((....)))))))).))...)))))).)))))..))))--). ( -45.60) >DroMoj_CAF1 61566 119 - 1 GGCUUGUUGAGAUGCCGACGCAGCCGCGACACAGUGGUCAGAUAUAUGGAAAUGUUUAGUAUGGUCAGCAUUGCGGCCACGAAGAUGGCCAGCGAGACAAAGGCCGUCAUAUCUGA-AGA (((((((((......))))).))))............((((((((((((...(((((.((.((((((...((.((....))))..)))))))).)))))....)))).))))))))-... ( -37.80) >DroAna_CAF1 52606 120 - 1 GGUUUGCUCAAGUGGCGGCACAUCCUCGAGACGGUGGUGGAGAAGAUGCAGAUGUUCAGGAUGGUCAGCAAACUGGCCACAAAAAUGGCCAAGCUGACAAAGGCAGUCAUGUCUGAUGAU .......(((.(((((.((.((((((.(((.((...(((.......)))...)))))))))))((((((....((((((......)))))).))))))....)).)))))...))).... ( -39.00) >DroPer_CAF1 59385 118 - 1 GGCUUGUUAAGGUGGCGCCUCAGACGCGACACAGUGGUCGAGUAGAUGCAGAUGUUGAGGAUGGUCAGCAGGCUGGCUAUGAAAAUGGCCAGGCUAAGGAACGCAGUCAUAUCUGA--GG .((((....))))....((((((((.((((......)))).)).(((((....(((((......)))))((.((((((((....)))))))).)).......))).))....))))--)) ( -42.00) >consensus GGCUUGUUCAGGUGGCGACGCAGCCGCGACACAGUGGUCGAGUAGAUGCAGAUGUUGAGGAUGGUCAGCAGGCUGGCUAUGAAAAUGGCCAGGCUGAGAAAGGCAGUCAUAUCUGA_AGA ...........(((((....................................((((((......))))))((((((((((....)))))).))))..........))))).......... (-20.52 = -19.63 + -0.89)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:35:17 2006