
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 18,862,104 – 18,862,203 |
| Length | 99 |
| Max. P | 0.983975 |

| Location | 18,862,104 – 18,862,203 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 76.99 |
| Mean single sequence MFE | -25.58 |
| Consensus MFE | -14.74 |
| Energy contribution | -14.86 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.889802 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
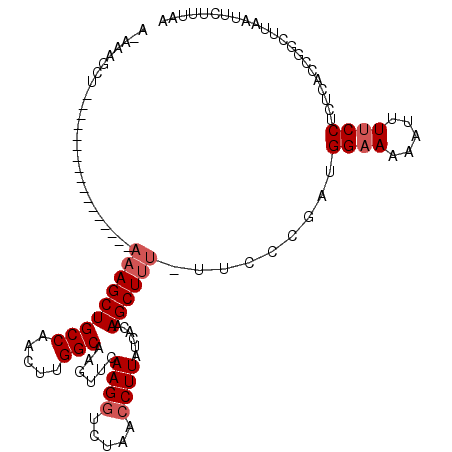
>3L_DroMel_CAF1 18862104 99 + 23771897 UUAAAGAAUUAAGCAGGUGGGAGGAAAAUUUUUCCAUCGGGAAAAAAGCUUGUUAUAAGGUUAGACCUUGAACUUGCCAAGCUGGCAGCUUU-----------------AGCUCU-U ...........(((.((((((((((...))))))))))......(((((..(((.(((((.....)))))))).((((.....)))))))))-----------------.)))..-. ( -27.40) >DroSec_CAF1 27047 98 + 1 UUAAAGUAUUAAGCCAGUGAGAGGAAAAAUUUUCCAUCGGGAA-AAAGCUUGUGAUAAGGUUCGAUCUUGAACUUGCCAAGUUGGCAGCUUU-----------------AGCUUU-U (((((((.....(((..............((((((....))))-))((((((.(...(((((((....))))))).)))))))))).)))))-----------------))....-. ( -23.50) >DroSim_CAF1 27593 98 + 1 UUAAAGAAUUAAGCCGCUGAGAGGAAAAAUUUUCCAUCGGGAA-AAAGCUUGUGAUAAGGUUAGACCUUGAACUUGCCAAGUUGGCAGCUUU-----------------AGCUUU-U ..........((((..((((..((((.....)))).))))...-(((((..((..(((((.....))))).)).((((.....)))))))))-----------------.)))).-. ( -24.10) >DroYak_CAF1 28072 92 + 1 UUAAAGAAUUAAAAAGGUGAGAGGAAAAUUUUUCCUUUAGGAA-AAAGCUUGUGAUAAGGUUAGACCUUGAACUUGCCAAUCUGGCAGCU-----------------------UU-U ...........(((((.((...(((...(((((((....))))-)))((..((..(((((.....))))).))..))...)))..)).))-----------------------))-) ( -21.50) >DroAna_CAF1 27384 113 + 1 AAAUCUAUUUAGAUUAAUGAGGGGAAAAUUUUACCCUCAG----AAAGCUUGUGAUAAGGUCAGACCUUGAACUUGCCAAGUUGGCAGCUUUUUGGCGUAAAACUAAAAAGCUUUAU .(((((....)))))..((((((..........))))))(----..((((((.(.((((.((((...)))).)))))))))))..)((((((((((.......)))))))))).... ( -31.40) >consensus UUAAAGAAUUAAGCAGGUGAGAGGAAAAUUUUUCCAUCGGGAA_AAAGCUUGUGAUAAGGUUAGACCUUGAACUUGCCAAGUUGGCAGCUUU_________________AGCUUU_U ....................((((((.....)))).))......(((((......(((((.....)))))....((((.....)))))))))......................... (-14.74 = -14.86 + 0.12)


| Location | 18,862,104 – 18,862,203 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 76.99 |
| Mean single sequence MFE | -21.08 |
| Consensus MFE | -12.18 |
| Energy contribution | -12.98 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.58 |
| SVM decision value | 1.96 |
| SVM RNA-class probability | 0.983975 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18862104 99 - 23771897 A-AGAGCU-----------------AAAGCUGCCAGCUUGGCAAGUUCAAGGUCUAACCUUAUAACAAGCUUUUUUCCCGAUGGAAAAAUUUUCCUCCCACCUGCUUAAUUCUUUAA .-.((((.-----------------.((((((((.....)))..(((.((((.....))))..))).)))))((((((....))))))...............)))).......... ( -20.80) >DroSec_CAF1 27047 98 - 1 A-AAAGCU-----------------AAAGCUGCCAACUUGGCAAGUUCAAGAUCGAACCUUAUCACAAGCUUU-UUCCCGAUGGAAAAUUUUUCCUCUCACUGGCUUAAUACUUUAA .-.(((((-----------------(((((((((.....)))..((((......)))).........))))).-.....((.((((.....))))))....)))))).......... ( -20.80) >DroSim_CAF1 27593 98 - 1 A-AAAGCU-----------------AAAGCUGCCAACUUGGCAAGUUCAAGGUCUAACCUUAUCACAAGCUUU-UUCCCGAUGGAAAAUUUUUCCUCUCAGCGGCUUAAUUCUUUAA .-.(((((-----------------(((((((((.....)))......((((.....))))......))))))-.....((.((((.....)))))).....))))).......... ( -21.20) >DroYak_CAF1 28072 92 - 1 A-AA-----------------------AGCUGCCAGAUUGGCAAGUUCAAGGUCUAACCUUAUCACAAGCUUU-UUCCUAAAGGAAAAAUUUUCCUCUCACCUUUUUAAUUCUUUAA (-((-----------------------(((((((.....)))......((((.....))))......))))))-)......(((((.....)))))..................... ( -16.80) >DroAna_CAF1 27384 113 - 1 AUAAAGCUUUUUAGUUUUACGCCAAAAAGCUGCCAACUUGGCAAGUUCAAGGUCUGACCUUAUCACAAGCUUU----CUGAGGGUAAAAUUUUCCCCUCAUUAAUCUAAAUAGAUUU .(((((((....)))))))......(((((((((.....)))......((((.....))))......))))))----.((((((..........))))))..(((((....))))). ( -25.80) >consensus A_AAAGCU_________________AAAGCUGCCAACUUGGCAAGUUCAAGGUCUAACCUUAUCACAAGCUUU_UUCCCGAUGGAAAAAUUUUCCUCUCACCGGCUUAAUUCUUUAA .........................(((((((((.....)))......((((.....))))......)))))).........((((.....))))...................... (-12.18 = -12.98 + 0.80)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:35:04 2006