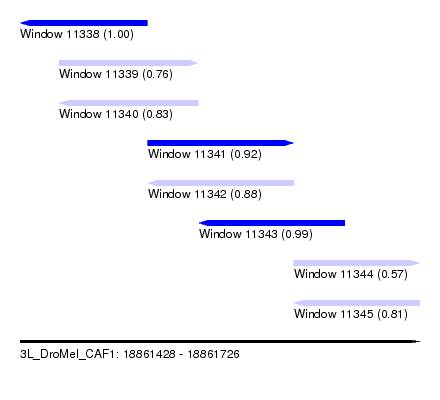
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 18,861,428 – 18,861,726 |
| Length | 298 |
| Max. P | 0.999777 |

| Location | 18,861,428 – 18,861,523 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 68.46 |
| Mean single sequence MFE | -29.22 |
| Consensus MFE | -11.51 |
| Energy contribution | -13.78 |
| Covariance contribution | 2.27 |
| Combinations/Pair | 1.38 |
| Mean z-score | -3.45 |
| Structure conservation index | 0.39 |
| SVM decision value | 4.06 |
| SVM RNA-class probability | 0.999777 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18861428 95 - 23771897 AGUGGGGGGAGGACGACGACUUGUUGUUGUUGUCCCCCUG--CAAACCAUCUAAUUCCUAGUUGGGAAGGUUACAUUU-CCU-UUUUA---UCACCGAUCAC .(..((((((.((((((((....)))))))).))))))..--)...(((.(((.....))).)))(((((........-)))-))...---........... ( -37.30) >DroSec_CAF1 26371 95 - 1 AGUGGGCGGAGAAGAACGACUUGUUGUUGUUCUCCCCCUG--CAAGCCAUCUAAUUCCUAGUUGAGAAGGUUACAUUU-CCU-UUUUA---UCACCUAACAC .(..((.((((((.(((((....))))).)))))).))..--).................(.((((((((........-)))-)))))---.)......... ( -29.60) >DroSim_CAF1 26923 95 - 1 AGUGGGGGGAGAAGAACGACUUGUUGUUGUUCUCCCCCUG--CAAGCCAUCUAAUUCCUAGUUGGGAAGGUUACAUUU-CCU-UUUUA---UCACCUAACAC .(..(((((((((.(((((....))))).)))))))))..--).................((((((((((........-)))-)....---...)))))).. ( -37.91) >DroYak_CAF1 27446 77 - 1 AGCGGGAGG---------ACU---------CGUCCUCCUG--CCAACCAUCUAAUUCCUAGUUGGGAAGGUUACAUUU-CUU-UUUUA---UCACCCAUUAC .((((((((---------(..---------..))))))))--).((((.((((((.....))))))..))))......-...-.....---........... ( -22.90) >DroMoj_CAF1 25826 86 - 1 AGCAGGUGG----G----------CGUUAAUUUCCCCUUGUACAAAC-CGCUGGGUCCUAGUUGGAGAAUUGACAAAUUAUCUUUUUGUUUUUAUCCAUUA- .(((((.((----(----------.(.....).))))))))......-.(((((...)))))((((.....((((((.......))))))....))))...- ( -18.40) >consensus AGUGGGGGGAG_AG_ACGACUUGUUGUUGUUCUCCCCCUG__CAAACCAUCUAAUUCCUAGUUGGGAAGGUUACAUUU_CCU_UUUUA___UCACCCAUCAC ..(((((((...((((((((.....)))))))))))))))......(((.(((.....))).)))..................................... (-11.51 = -13.78 + 2.27)



| Location | 18,861,457 – 18,861,561 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 77.45 |
| Mean single sequence MFE | -31.10 |
| Consensus MFE | -18.29 |
| Energy contribution | -17.94 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.764640 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18861457 104 + 23771897 UUCCCAACUAGGAAUUAGAUGGUUUGCAGGGGGACAACAACAACAAGUCGUCGUCCUCCCCCCACUCAGGUAAUUCAACUCUCUAGGGAAGGGCGCCCGGCCAA (((((......((((((..(((..((..(((((((.((.((.....)).)).)))).)))..)).)))..)))))).........))))).(((.....))).. ( -30.16) >DroSec_CAF1 26400 104 + 1 UUCUCAACUAGGAAUUAGAUGGCUUGCAGGGGGAGAACAACAACAAGUCGUUCUUCUCCGCCCACUCAGGUAAUUCAACUCUCUAGGGAAGGGCGCCCGGCCAA ((((......)))).....(((((.((.((((((((((.((.....)).))))))))))((((((....)).......(((....)))..))))))..))))). ( -35.30) >DroSim_CAF1 26952 104 + 1 UUCCCAACUAGGAAUUAGAUGGCUUGCAGGGGGAGAACAACAACAAGUCGUUCUUCUCCCCCCACUCAGGUAAUUCAACUCUCUAGGGAAGGGCGCCCGGCCAA (((((...(((((.((..((.(((((..(((((((((.(((........))).)))))))))....))))).))..)).)).)))))))).(((.....))).. ( -37.40) >DroEre_CAF1 26945 82 + 1 UUCCCAACUAGGAAUUAGAUGGUUGGCAGGAGGACG---------AGU---------CCUCC----CAGGUAAUUCAACUCUCUAGGGAAGGGCGCCCGACCAA ((((......))))(((((.(((((((.((((((..---------..)---------)))))----...))....))))).)))))((..((....))..)).. ( -26.90) >DroYak_CAF1 27475 86 + 1 UUCCCAACUAGGAAUUAGAUGGUUGGCAGGAGGACG---------AGU---------CCUCCCGCUCAGGUAAUUCAACUCUCUAGGGAAGGGCGCCCGACCAA ((((......)))).....(((((((..((((((..---------..)---------)))))(((((.((....))..(((....)))..))))).))))))). ( -30.60) >DroAna_CAF1 26729 85 + 1 AUCCCAACUAGGAGCCAGUUGGAUUGCAGGAGAAGUACA-------------------CCCCCACUCAAUUGGCUCAACUCUCUAGGGAGGGGCGCCCGGCCAA .((((......(((((((((((..((..((.(.....).-------------------))..)).))))))))))).........))))..(((.....))).. ( -26.26) >consensus UUCCCAACUAGGAAUUAGAUGGUUUGCAGGAGGACAACA______AGU_________CCCCCCACUCAGGUAAUUCAACUCUCUAGGGAAGGGCGCCCGGCCAA (((((......((((((..(((......(((((........................)))))...)))..)))))).........)))))((.(....).)).. (-18.29 = -17.94 + -0.36)


| Location | 18,861,457 – 18,861,561 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 77.45 |
| Mean single sequence MFE | -33.59 |
| Consensus MFE | -21.20 |
| Energy contribution | -20.71 |
| Covariance contribution | -0.49 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.825673 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18861457 104 - 23771897 UUGGCCGGGCGCCCUUCCCUAGAGAGUUGAAUUACCUGAGUGGGGGGAGGACGACGACUUGUUGUUGUUGUCCCCCUGCAAACCAUCUAAUUCCUAGUUGGGAA ..(((.....)))....(((((..((..((((((..((.(..((((((.((((((((....)))))))).))))))..)....))..))))))))..))))).. ( -40.60) >DroSec_CAF1 26400 104 - 1 UUGGCCGGGCGCCCUUCCCUAGAGAGUUGAAUUACCUGAGUGGGCGGAGAAGAACGACUUGUUGUUGUUCUCCCCCUGCAAGCCAUCUAAUUCCUAGUUGAGAA ..(((.....)))(((..((((.((((((.((..(.((...(((.((((((.(((((....))))).)))))))))..)).)..)).))))))))))..))).. ( -35.60) >DroSim_CAF1 26952 104 - 1 UUGGCCGGGCGCCCUUCCCUAGAGAGUUGAAUUACCUGAGUGGGGGGAGAAGAACGACUUGUUGUUGUUCUCCCCCUGCAAGCCAUCUAAUUCCUAGUUGGGAA ..(((.....)))..(((((((.((((((.((...((..(..(((((((((.(((((....))))).)))))))))..).))..)).)))))))))...)))). ( -42.00) >DroEre_CAF1 26945 82 - 1 UUGGUCGGGCGCCCUUCCCUAGAGAGUUGAAUUACCUG----GGAGG---------ACU---------CGUCCUCCUGCCAACCAUCUAAUUCCUAGUUGGGAA .((((..(((((.(((.....))).))..........(----(((((---------(..---------..)))))))))).)))).....(((((....))))) ( -24.70) >DroYak_CAF1 27475 86 - 1 UUGGUCGGGCGCCCUUCCCUAGAGAGUUGAAUUACCUGAGCGGGAGG---------ACU---------CGUCCUCCUGCCAACCAUCUAAUUCCUAGUUGGGAA ......((....)).(((((((.((((((.......((.((((((((---------(..---------..)))))))))))......)))))))))...)))). ( -30.32) >DroAna_CAF1 26729 85 - 1 UUGGCCGGGCGCCCCUCCCUAGAGAGUUGAGCCAAUUGAGUGGGGG-------------------UGUACUUCUCCUGCAAUCCAACUGGCUCCUAGUUGGGAU ((((((((....))(((....)))....).)))))....(..((((-------------------.......))))..)..(((((((((...))))))))).. ( -28.30) >consensus UUGGCCGGGCGCCCUUCCCUAGAGAGUUGAAUUACCUGAGUGGGGGG_________ACU______UGUUCUCCCCCUGCAAACCAUCUAAUUCCUAGUUGGGAA ..(((.....)))....(((((..((..((((((..((.((((((((........................))))))))....))..))))))))..))))).. (-21.20 = -20.71 + -0.49)



| Location | 18,861,523 – 18,861,632 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 78.69 |
| Mean single sequence MFE | -46.83 |
| Consensus MFE | -24.76 |
| Energy contribution | -25.07 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.53 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.915578 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18861523 109 + 23771897 CAGGUAAUUCAACUCUCUAGGGAAGGGCGCCCGGCCAAGUGCC--ACGCCCCGGAUUGCCCG-GCGUGGCCACUAGAACAGUGGCCACAGGUGGACAGGGGGAAUGGCACAG ..(((.......(((....)))..((....)).)))..(((((--(..((((...(((.(((-.((((((((((.....))))))))).).))).)))))))..)))))).. ( -50.20) >DroVir_CAF1 27657 105 + 1 CCACUGACUCAACUCUCUAGGCGAAUU-ACGCGACCAAGUGCCUCGCACAUC-CGUUGCCCGCGCG---CCACUAGAACAGUGGACACAAU-GGGCGGA-GGCGUGGCACAG ....................(((....-.)))......(((((.(((...((-(...(((((.(.(---(((((.....))))).).)..)-)))))))-.))).))))).. ( -36.00) >DroEre_CAF1 26989 109 + 1 CAGGUAAUUCAACUCUCUAGGGAAGGGCGCCCGACCAAGUGCC--ACGCCCCGGAUUGUCCG-GCGUGAUCACUAGAACAGUGACCACAGGUGGGCAGGGGGCGUGGCACAG ..(((.......(((....)))..((....)).)))..(((((--(((((((...(((((((-.((((.(((((.....))))).))).).))))))))))))))))))).. ( -54.10) >DroYak_CAF1 27523 109 + 1 CAGGUAAUUCAACUCUCUAGGGAAGGGCGCCCGACCAAGUGCC--ACGCCCCGAAUUGUCCG-GCGUGAUCACUAGAACAGUGACCACAGGUGGGCAGGGGGCGUGGCACAG ..(((.......(((....)))..((....)).)))..(((((--(((((((...(((((((-.((((.(((((.....))))).))).).))))))))))))))))))).. ( -54.10) >DroMoj_CAF1 25912 102 + 1 CCGCUGGCUCAACUCUCUAGGCGAAUU-ACGCGACCAAGAACC--ACACAUC-CAUUGCCCGUGCG---CCACUAGAACAGUG-ACACAAU-GGGCGGA-GGCGUGGCACAG ...(((((.......(((.((((....-...)).)).)))...--.(((..(-(.(((((((((.(---.((((.....))))-.).).))-)))))).-)).))))).))) ( -28.60) >DroAna_CAF1 26776 109 + 1 CAAUUGGCUCAACUCUCUAGGGAGGGGCGCCCGGCCAAGUGCC--ACGCCCCCGAUUGCCCG-GCGCGAACACUAGAACAGUGACCACAGGUGGGCGGGGGGUGUGGCACAG ...((((((...((((....))))((....))))))))(((((--((((((((...((((((-.(.....((((.....))))......).))))))))))))))))))).. ( -58.00) >consensus CAGGUAACUCAACUCUCUAGGGAAGGGCGCCCGACCAAGUGCC__ACGCCCCGGAUUGCCCG_GCGUGACCACUAGAACAGUGACCACAGGUGGGCAGGGGGCGUGGCACAG ...................((...((....))..))..(((((...(.((((....((((((.......(((((.....))))).......)))))))))))...))))).. (-24.76 = -25.07 + 0.31)


| Location | 18,861,523 – 18,861,632 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 78.69 |
| Mean single sequence MFE | -45.67 |
| Consensus MFE | -30.53 |
| Energy contribution | -31.78 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.881876 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18861523 109 - 23771897 CUGUGCCAUUCCCCCUGUCCACCUGUGGCCACUGUUCUAGUGGCCACGC-CGGGCAAUCCGGGGCGU--GGCACUUGGCCGGGCGCCCUUCCCUAGAGAGUUGAAUUACCUG ..(((((((.((((.(((((...(((((((((((...))))))))))).-.)))))....)))).))--)))))..((..((....))..)).................... ( -47.30) >DroVir_CAF1 27657 105 - 1 CUGUGCCACGCC-UCCGCCC-AUUGUGUCCACUGUUCUAGUGG---CGCGCGGGCAACG-GAUGUGCGAGGCACUUGGUCGCGU-AAUUCGCCUAGAGAGUUGAGUCAGUGG ..(((((.((((-(((((((-..((((.((((((...))))))---)))).))))...)-)).).))).))))).....(((.(-(((((.......)))))).....))). ( -39.70) >DroEre_CAF1 26989 109 - 1 CUGUGCCACGCCCCCUGCCCACCUGUGGUCACUGUUCUAGUGAUCACGC-CGGACAAUCCGGGGCGU--GGCACUUGGUCGGGCGCCCUUCCCUAGAGAGUUGAAUUACCUG ..((((((((((((..........((((((((((...))))))))))..-.((.....)))))))))--)))))..((..((....))..)).................... ( -47.30) >DroYak_CAF1 27523 109 - 1 CUGUGCCACGCCCCCUGCCCACCUGUGGUCACUGUUCUAGUGAUCACGC-CGGACAAUUCGGGGCGU--GGCACUUGGUCGGGCGCCCUUCCCUAGAGAGUUGAAUUACCUG ..((((((((((((.((.((...(((((((((((...))))))))))).-.)).))....)))))))--)))))..((..((....))..)).................... ( -46.70) >DroMoj_CAF1 25912 102 - 1 CUGUGCCACGCC-UCCGCCC-AUUGUGU-CACUGUUCUAGUGG---CGCACGGGCAAUG-GAUGUGU--GGUUCUUGGUCGCGU-AAUUCGCCUAGAGAGUUGAGCCAGCGG (((((((((((.-(((((((-..(((((-(((((...))))))---)))).))))...)-)).))))--)))...((((....(-(((((.......)))))).)))))))) ( -42.70) >DroAna_CAF1 26776 109 - 1 CUGUGCCACACCCCCCGCCCACCUGUGGUCACUGUUCUAGUGUUCGCGC-CGGGCAAUCGGGGGCGU--GGCACUUGGCCGGGCGCCCCUCCCUAGAGAGUUGAGCCAAUUG ..(((((((.(((((.((((...((((..(((((...)))))..)))).-.))))....))))).))--)))))((((((((....))(((....)))....).)))))... ( -50.30) >consensus CUGUGCCACGCCCCCCGCCCACCUGUGGUCACUGUUCUAGUGGUCACGC_CGGGCAAUCCGGGGCGU__GGCACUUGGUCGGGCGCCCUUCCCUAGAGAGUUGAAUCACCUG ..((((((((((((..((((...(((((((((((...)))))))))))...)))).....)))))))..)))))((((..(((......)))))))................ (-30.53 = -31.78 + 1.25)



| Location | 18,861,561 – 18,861,670 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.13 |
| Mean single sequence MFE | -44.82 |
| Consensus MFE | -22.64 |
| Energy contribution | -23.03 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.39 |
| Mean z-score | -2.81 |
| Structure conservation index | 0.51 |
| SVM decision value | 2.23 |
| SVM RNA-class probability | 0.990778 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18861561 109 - 23771897 CGAAUAGCGUUACACCAGUGAUAGAGCAAUA--AAUGCGGC-UGUGCCAU--UCCCCCUGUCCACCUGUGGCCACUGUUCUAGUGGCCACGC-CGGGCAAUCCGG-GGCGU----GGCAC .....((((((((....)))))...(((...--..))).))-)(((((((--.((((.(((((...(((((((((((...))))))))))).-.)))))....))-)).))----))))) ( -50.00) >DroVir_CAF1 27694 106 - 1 UCCACAGCGUUACACUUGUGAUAGAGCAAAU--AAUGCGGC-UGUGCCAC--GCC-UCCGCCC-AUUGUGUCCACUGUUCUAGUGG---CGCGCGGGCAACG-GA-UGUGC--GAGGCAC ...((((((((((....)))))...(((...--..))).))-)))(((.(--(((-(((((((-..((((.((((((...))))))---)))).))))...)-))-.).))--).))).. ( -42.60) >DroGri_CAF1 32645 119 - 1 UCUACAGCGUUACACUUGUGAUAGAGCAAUUAAAAUGCGACUUGUGGCAAAAGCCGGCCGCUCAACUGUGUUCACUGUUCUAGUGGAAACGACGGGGCAAUG-GAAUGUGCUUUAGUCAC ......(((((....((((......))))....)))))((((...((((....(((...((((...(((.(((((((...))))))).)))...))))..))-)....))))..)))).. ( -33.50) >DroYak_CAF1 27561 109 - 1 CGAAUAGCGUUACACCUGUGAUAGAGCAAUA--AAUGCGGC-UGUGCCAC--GCCCCCUGCCCACCUGUGGUCACUGUUCUAGUGAUCACGC-CGGACAAUUCGG-GGCGU----GGCAC .....((((((((....)))))...(((...--..))).))-)(((((((--(((((.((.((...(((((((((((...))))))))))).-.)).))....))-)))))----))))) ( -48.90) >DroMoj_CAF1 25949 102 - 1 UCCACAGCGUUACACUUGUGAUAGAGCAAAU---AUGCGGC-UGUGCCAC--GCC-UCCGCCC-AUUGUGU-CACUGUUCUAGUGG---CGCACGGGCAAUG-GA-UGUGU----GGUUC ..(((((((((((....)))))...(((...---.))).))-))))((((--((.-(((((((-..(((((-(((((...))))))---)))).))))...)-))-.))))----))... ( -46.30) >DroAna_CAF1 26814 109 - 1 CAAAAGGCGUUACACUUGUGAUAGAGCAUUA--AAUGCGGC-UGUGCCAC--ACCCCCCGCCCACCUGUGGUCACUGUUCUAGUGUUCGCGC-CGGGCAAUCGGG-GGCGU----GGCAC .....((((((((....)))))...((((..--.)))).))-)(((((((--.(((((.((((...((((..(((((...)))))..)))).-.))))....)))-)).))----))))) ( -47.60) >consensus CCAACAGCGUUACACUUGUGAUAGAGCAAUA__AAUGCGGC_UGUGCCAC__GCCCCCCGCCCAACUGUGGUCACUGUUCUAGUGG_CACGC_CGGGCAAUG_GA_GGCGU____GGCAC ......(((((.....(((......))).....))))).....(((((....((((((.((((...(((.(((((((...))))))).)))...)))).....)).)).))....))))) (-22.64 = -23.03 + 0.39)

| Location | 18,861,632 – 18,861,726 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 80.03 |
| Mean single sequence MFE | -22.07 |
| Consensus MFE | -18.68 |
| Energy contribution | -18.38 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.08 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.567628 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18861632 94 + 23771897 CCGCAUU--UAUUGCUCUAUCACUGGUGUAACGCUAUUCGGUUGACGUUUAAAACCAGAACCGUACAGCUGACUGACCUGUGAAAAC---UC-GUG-GUUA (((((..--...)))....((((.(((((...(((...(((((...((.....))...)))))...)))..))..))).))))....---..-..)-)... ( -19.40) >DroVir_CAF1 27762 96 + 1 CCGCAUU--AUUUGCUCUAUCACAAGUGUAACGCUGUGGAGUUCAUGU---AAAUUACAACUGAACAGCUGACUGACUUGUGAACAGAUCCCAAAUUGAUA ..(((..--...)))(((.((((((((((...(((((..(((...(((---(...)))))))..)))))..))..))))))))..)))............. ( -22.70) >DroGri_CAF1 32724 95 + 1 UCGCAUUUUAAUUGCUCUAUCACAAGUGUAACGCUGUAGAGUUAAAUU--CAAGUAACAACUAAACAGCUGACUGACUUGUGAAAAC---CC-AAGCGAAA ((((.......(((.....((((((((((...(((((..((((.....--........))))..)))))..))..))))))))....---.)-)))))).. ( -21.03) >DroSec_CAF1 26575 94 + 1 CCGCAUU--UAUUGCUCUAUCACAGGUGUAACGCUAUUCGGUUGACGUGUAAAACCAGAACCGUACAGCUGACUGACCUGUGAAAAC---AC-GUG-GUUA (((((..--...)))....((((((((((...(((...(((((...((.....))...)))))...)))..))..))))))))....---..-..)-)... ( -25.10) >DroSim_CAF1 27127 94 + 1 CCGCAUU--UAUUGCUCUAUCACUGGUGUAACGCUAUUCGGUUGACGUGUAAAACCAGAACCGUACAGCUGACUGACCUGUGAAAAC---AC-GUG-GUUA ..(((..--...)))........(((((...))))).((((((((((.((.........))))).))))))).(((((((((....)---))-).)-)))) ( -21.20) >DroEre_CAF1 27098 90 + 1 CCGCAUU--UAUUGCUCUAUCACAGGUGUAACGCUAUUCGGUUGACGUUUAAAACCAGAACCGUACAGCUGACUGACCUGUGAAAA-----C-GUA-G--A ..(((..--...)))....((((((((((...(((...(((((...((.....))...)))))...)))..))..))))))))...-----.-...-.--. ( -23.00) >consensus CCGCAUU__UAUUGCUCUAUCACAGGUGUAACGCUAUUCGGUUGACGU_UAAAACCAGAACCGUACAGCUGACUGACCUGUGAAAAC___AC_GUG_GUUA ..(((.......)))....((((((((((...(((...(((((...((.....))...)))))...)))..))..)))))))).................. (-18.68 = -18.38 + -0.30)


| Location | 18,861,632 – 18,861,726 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 80.03 |
| Mean single sequence MFE | -24.92 |
| Consensus MFE | -22.15 |
| Energy contribution | -21.71 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.814352 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18861632 94 - 23771897 UAAC-CAC-GA---GUUUUCACAGGUCAGUCAGCUGUACGGUUCUGGUUUUAAACGUCAACCGAAUAGCGUUACACCAGUGAUAGAGCAAUA--AAUGCGG ...(-(..-..---....((((.(((......(((((.(((((.((........))..))))).))))).....))).))))....(((...--..))))) ( -22.40) >DroVir_CAF1 27762 96 - 1 UAUCAAUUUGGGAUCUGUUCACAAGUCAGUCAGCUGUUCAGUUGUAAUUU---ACAUGAACUCCACAGCGUUACACUUGUGAUAGAGCAAAU--AAUGCGG .....(((((...(((((.(((((((......(((((..(((((((...)---)))...)))..))))).....)))))))))))).)))))--....... ( -24.10) >DroGri_CAF1 32724 95 - 1 UUUCGCUU-GG---GUUUUCACAAGUCAGUCAGCUGUUUAGUUGUUACUUG--AAUUUAACUCUACAGCGUUACACUUGUGAUAGAGCAAUUAAAAUGCGA ..((((..-..---((((((((((((......(((((..(((((.......--....)))))..))))).....))))))))..)))).........)))) ( -25.22) >DroSec_CAF1 26575 94 - 1 UAAC-CAC-GU---GUUUUCACAGGUCAGUCAGCUGUACGGUUCUGGUUUUACACGUCAACCGAAUAGCGUUACACCUGUGAUAGAGCAAUA--AAUGCGG ....-..(-((---((((((((((((......(((((.(((((.((........))..))))).))))).....)))))))).........)--)))))). ( -27.90) >DroSim_CAF1 27127 94 - 1 UAAC-CAC-GU---GUUUUCACAGGUCAGUCAGCUGUACGGUUCUGGUUUUACACGUCAACCGAAUAGCGUUACACCAGUGAUAGAGCAAUA--AAUGCGG ....-..(-((---((((((((.(((......(((((.(((((.((........))..))))).))))).....))).)))).........)--)))))). ( -22.60) >DroEre_CAF1 27098 90 - 1 U--C-UAC-G-----UUUUCACAGGUCAGUCAGCUGUACGGUUCUGGUUUUAAACGUCAACCGAAUAGCGUUACACCUGUGAUAGAGCAAUA--AAUGCGG (--(-((.-.-----...((((((((......(((((.(((((.((........))..))))).))))).....))))))))))))(((...--..))).. ( -27.30) >consensus UAAC_CAC_GG___GUUUUCACAGGUCAGUCAGCUGUACGGUUCUGGUUUUA_ACGUCAACCGAAUAGCGUUACACCUGUGAUAGAGCAAUA__AAUGCGG ..................((((((((......(((((.(((((...............))))).))))).....))))))))....(((.......))).. (-22.15 = -21.71 + -0.44)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:35:02 2006