
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 18,850,899 – 18,851,132 |
| Length | 233 |
| Max. P | 0.999262 |

| Location | 18,850,899 – 18,851,000 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 71.26 |
| Mean single sequence MFE | -26.65 |
| Consensus MFE | -14.19 |
| Energy contribution | -14.50 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.53 |
| SVM decision value | 1.34 |
| SVM RNA-class probability | 0.943565 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18850899 101 + 23771897 UAGCAAUUUCAGUGCUCAUACACA-CUUAUGUAUAUUGGAAAACACUCUG-----UGAAGA------AGAAAGAGUGUGCCAAAUG----UUGGUGCAUUCUAAUCUGUACGCCAUA ..((..((((((((..((((....-..))))..))))))))........(-----((.(((------....((((((..(((....----.)))..))))))..))).))))).... ( -26.50) >DroSec_CAF1 16061 97 + 1 UAGCAAUUUUAGUGCGCAUACACA-CUUAU----GUUGGAAAACACUCUG-----UGAAAGG---CGAAAAAGAGUGUGCCAAAUG----UUGGUGCAUUCUAGUCUA---GUCAUA ...((((...((((.(....).))-))...----))))..........((-----(((.(((---(.....((((((..(((....----.)))..)))))).)))).---.))))) ( -25.70) >DroSim_CAF1 16008 95 + 1 UAGCAAUUUUAGUGCUCAUACACA-CUUAU----AUUGGAAAACACUCUG-----UGAAAAG-----AGAAAGAGUGUGCCAAAUG----UUGGUGCAUUCUAGUCUA---GUCAUA ...((((...((((........))-))...----))))..........((-----(((..((-----(...((((((..(((....----.)))..))))))..))).---.))))) ( -24.90) >DroEre_CAF1 16467 102 + 1 UAGCAAUUUUGGUGUUCGUACACA-CUUAU----AUUGGAAAACACUCUACACUUUGUAGG------GGAAAGAGUGUGCCAAAUG----UUGGUGCAUUCGAGUUUGUAUGCCAGA .......((((((....(((((.(-(((..----...........((((((.....)))))------)....(((((..(((....----.)))..))))))))).))))))))))) ( -32.30) >DroYak_CAF1 16782 102 + 1 UAGCAAUUUUAGUGUCCACACACA-CUUGU----AUUGGAAAACACUCUACUCAAUGUAGG------AGAAAGAGUGUGCCAAAUG----CUGGUGCAUUCGAACUUGCCCGCCAGA ..((((.(((.(((....))).((-((.((----(((((...(((((((.(((.......)------))..))))))).)).))))----).)))).....))).))))........ ( -28.30) >DroAna_CAF1 17597 98 + 1 UAGCCGCCUUAGUGUUCAUGCGAGUAGUCC----AUUGAAAAGCCGUCUA-----UAAACGCAUUUGGUUAAGAGUGUGCCAAAUGCUUGUUGGUGCAGU----------UGCUAGA ((((.((.((((((..(.((....)))..)----)))))...(((((...-----...))(((((((((.........))))))))).....)))...))----------.)))).. ( -22.20) >consensus UAGCAAUUUUAGUGCUCAUACACA_CUUAU____AUUGGAAAACACUCUA_____UGAAGG______AGAAAGAGUGUGCCAAAUG____UUGGUGCAUUCUAGUCUG___GCCAGA ..((.......(((......))).................................................(((((..((((.......))))..)))))..........)).... (-14.19 = -14.50 + 0.31)



| Location | 18,850,938 – 18,851,040 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 72.55 |
| Mean single sequence MFE | -29.82 |
| Consensus MFE | -13.94 |
| Energy contribution | -15.22 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.48 |
| Structure conservation index | 0.47 |
| SVM decision value | 3.47 |
| SVM RNA-class probability | 0.999262 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18850938 102 + 23771897 AAACACUCUG-----UGAAGA------AGAAAGAGUGUGCCAAAUG----UUGGUGCAUUCUAAUCUGUACGCCAUAGCAGACUAAACAAGCACACUAAGUGAGUGAAAUUCCGAAA ...(((((.(-----((....------....((((((..(((....----.)))..))))))..(((((........)))))...........))).....)))))........... ( -27.50) >DroSec_CAF1 16096 102 + 1 AAACACUCUG-----UGAAAGG---CGAAAAAGAGUGUGCCAAAUG----UUGGUGCAUUCUAGUCUA---GUCAUACCAGACUAAACAAACACAAUAAGUGAGUGAAAUUCAGAAA ...(((((((-----((.....---......((((((..(((....----.)))..)))))).((.((---(((......))))).))...))))......)))))........... ( -29.50) >DroSim_CAF1 16043 100 + 1 AAACACUCUG-----UGAAAAG-----AGAAAGAGUGUGCCAAAUG----UUGGUGCAUUCUAGUCUA---GUCAUAACAGACUAAACAAACACACUAAGUGAGUGAAAUUCAGAAA ...(((((.(-----((.....-----....((((((..(((....----.)))..)))))).((.((---(((......))))).)).....))).....)))))........... ( -29.70) >DroEre_CAF1 16502 104 + 1 AAACACUCUACACUUUGUAGG------GGAAAGAGUGUGCCAAAUG----UUGGUGCAUUCGAGUUUGUAUGCCAGACCAGACUAGGCAAACACACUAAGUGAGUGAAAUUCAG--- .....((((((.....)))))------)....(((((..(((....----.)))..)))))((((((((.((((((......)).)))).)).((((.....))))))))))..--- ( -34.90) >DroYak_CAF1 16817 107 + 1 AAACACUCUACUCAAUGUAGG------AGAAAGAGUGUGCCAAAUG----CUGGUGCAUUCGAACUUGCCCGCCAGACCAGACUAGGCAAACACACCAAGUGAGUGAAAUUCAGAAA ...(((((.(((....(((((------.....(((((..(((....----.)))..)))))...)))))..(((((......)).)))..........))))))))........... ( -29.60) >DroAna_CAF1 17633 96 + 1 AAGCCGUCUA-----UAAACGCAUUUGGUUAAGAGUGUGCCAAAUGCUUGUUGGUGCAGU----------UGCUAGACCAGUCUGAACAUUCAGAAUUA-----UGCAGUUCAGA-A .....(((((-----..((((((((((((.........))))))))).(((....)))))----------)..)))))...(((((((...((......-----))..)))))))-. ( -27.70) >consensus AAACACUCUA_____UGAAGG______AGAAAGAGUGUGCCAAAUG____UUGGUGCAUUCUAGUCUG___GCCAGACCAGACUAAACAAACACACUAAGUGAGUGAAAUUCAGAAA ...(((((........................(((((..((((.......))))..)))))..........((.............)).............)))))........... (-13.94 = -15.22 + 1.28)



| Location | 18,850,938 – 18,851,040 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 72.55 |
| Mean single sequence MFE | -26.54 |
| Consensus MFE | -8.72 |
| Energy contribution | -9.80 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.33 |
| SVM decision value | 1.57 |
| SVM RNA-class probability | 0.964527 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18850938 102 - 23771897 UUUCGGAAUUUCACUCACUUAGUGUGCUUGUUUAGUCUGCUAUGGCGUACAGAUUAGAAUGCACCAA----CAUUUGGCACACUCUUUCU------UCUUCA-----CAGAGUGUUU ....((.....((((.....))))(((...(((((((((.(((...))))))))))))..)))))..----........(((((((....------......-----.))))))).. ( -21.50) >DroSec_CAF1 16096 102 - 1 UUUCUGAAUUUCACUCACUUAUUGUGUUUGUUUAGUCUGGUAUGAC---UAGACUAGAAUGCACCAA----CAUUUGGCACACUCUUUUUCG---CCUUUCA-----CAGAGUGUUU ...........(((((.......((((...((((((((((.....)---)))))))))..))))...----.....(((............)---)).....-----..)))))... ( -26.60) >DroSim_CAF1 16043 100 - 1 UUUCUGAAUUUCACUCACUUAGUGUGUUUGUUUAGUCUGUUAUGAC---UAGACUAGAAUGCACCAA----CAUUUGGCACACUCUUUCU-----CUUUUCA-----CAGAGUGUUU .(((((..............((((((((.((((((((......)))---)))))..(((((......----)))))))))))))......-----.......-----)))))..... ( -23.13) >DroEre_CAF1 16502 104 - 1 ---CUGAAUUUCACUCACUUAGUGUGUUUGCCUAGUCUGGUCUGGCAUACAAACUCGAAUGCACCAA----CAUUUGGCACACUCUUUCC------CCUACAAAGUGUAGAGUGUUU ---........(((((....(((((((.((((.((......))))))........((((((......----)))))))))))))......------..(((.....))))))))... ( -24.80) >DroYak_CAF1 16817 107 - 1 UUUCUGAAUUUCACUCACUUGGUGUGUUUGCCUAGUCUGGUCUGGCGGGCAAGUUCGAAUGCACCAG----CAUUUGGCACACUCUUUCU------CCUACAUUGAGUAGAGUGUUU ...........((((((((..(((((.((((((.(((......)))))))))((((((((((....)----))))))).)).........------..)))))..))).)))))... ( -37.00) >DroAna_CAF1 17633 96 - 1 U-UCUGAACUGCA-----UAAUUCUGAAUGUUCAGACUGGUCUAGCA----------ACUGCACCAACAAGCAUUUGGCACACUCUUAACCAAAUGCGUUUA-----UAGACGGCUU .-(((((((..((-----......))...)))))))...(((((((.----------...))........((((((((...........)))))))).....-----)))))..... ( -26.20) >consensus UUUCUGAAUUUCACUCACUUAGUGUGUUUGUUUAGUCUGGUAUGGC___CAGACUAGAAUGCACCAA____CAUUUGGCACACUCUUUCU______CCUUCA_____CAGAGUGUUU ...((((((..(((.........)))...))))))............................((((.......)))).(((((((......................))))))).. ( -8.72 = -9.80 + 1.08)


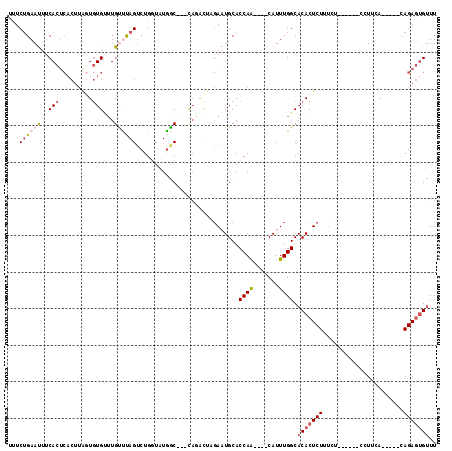
| Location | 18,851,000 – 18,851,120 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.82 |
| Mean single sequence MFE | -33.08 |
| Consensus MFE | -19.42 |
| Energy contribution | -20.76 |
| Covariance contribution | 1.34 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.59 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.914319 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18851000 120 - 23771897 AGACAGGGCCAACCUGUCUACAUGUGAGCAGAUACAUUUGCUUGUGCGGUUGUAAAACAAGUAUUUUGUAUAAUUAUGCAUUUCGGAAUUUCACUCACUUAGUGUGCUUGUUUAGUCUGC (((((((.....)))))))........(((((((((.((((....)))).))).(((((((((...(((((....)))))...........((((.....))))))))))))).)))))) ( -35.60) >DroSec_CAF1 16158 120 - 1 AGACAGGGCCAACCUGUCUACAUCUGAGCAGAUACAUUUGCGUGUGCGGUUGUAAAACAAGUAUUUUGUAUAAUUAUGCAUUUCUGAAUUUCACUCACUUAUUGUGUUUGUUUAGUCUGG (((((((.....))))))).((.(((((((((((((..((.(((.(.((((....)))........(((((....)))))...........).).))).)).)))))))))))))..)). ( -32.30) >DroSim_CAF1 16103 120 - 1 AGACAGGGCCAACCUGUCUACAUGUGAGCAGAUACAUUUGCGUGUGCGGUUGUAAAACAAGUAUUUUGUAUAAUUAUGCAUUUCUGAAUUUCACUCACUUAGUGUGUUUGUUUAGUCUGU (((((((.....))))))).....((((((((((((((...(((.(.((((....)))........(((((....)))))...........).).)))..))))))))))))))...... ( -31.80) >DroEre_CAF1 16569 117 - 1 AGACAGGGCCAACCUGUCUACAUGAGAGCAGAUACAUUUGCGUGUGCGGUUGUAAAACAAGUAUAAUAUAUAAUUAUGCA---CUGAAUUUCACUCACUUAGUGUGUUUGCCUAGUCUGG (((((((.....))))))).....((.(((((((((((((.(((..((((((.....)).(((((((.....))))))))---))).....))).))...)))))))))))))....... ( -34.40) >DroYak_CAF1 16884 120 - 1 AGACAGGGCCAACCUGUCUACAAGUGAGCAGAUACAUUUGAGAGUGCGGUUGUAAAACAAUGAUUGUGUAUAAUUAUGCAUUUCUGAAUUUCACUCACUUGGUGUGUUUGCCUAGUCUGG (((((((.....))))))).((..(..(((((((((((.(.(((((..((((.....))))((..((((((....)))))).)).......))))).)..)))))))))))...)..)). ( -36.50) >DroAna_CAF1 17695 97 - 1 UGACAGGGACAACCUGUCUGCUUAUGACCGGGUAUAGAUAUG--UAC-------AAAUAAAAAUAAU--------GUGCAU-UCUGAACUGCA-----UAAUUCUGAAUGUUCAGACUGG .((((((.....)))))).........((((........(((--(((-------(...........)--------))))))-(((((((..((-----......))...))))))))))) ( -27.90) >consensus AGACAGGGCCAACCUGUCUACAUGUGAGCAGAUACAUUUGCGUGUGCGGUUGUAAAACAAGUAUUUUGUAUAAUUAUGCAUUUCUGAAUUUCACUCACUUAGUGUGUUUGUUUAGUCUGG (((((((.....))))))).....((((((((((((((.....(((..((((((..............))))))..)))......(.....)........))))))))))))))...... (-19.42 = -20.76 + 1.34)

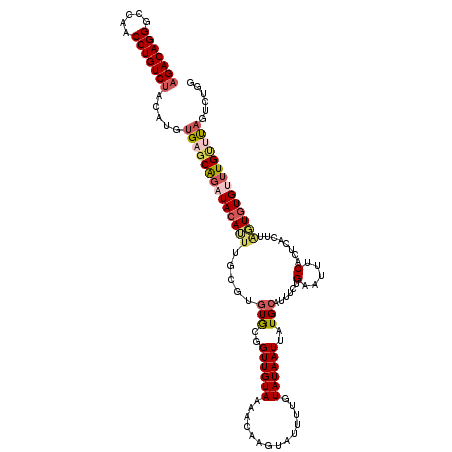

| Location | 18,851,040 – 18,851,132 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 80.71 |
| Mean single sequence MFE | -21.92 |
| Consensus MFE | -13.54 |
| Energy contribution | -14.90 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.583665 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18851040 92 - 23771897 AGACUU-------UUGACAAGACAGGGCCAACCUGUCUACAUGUGAGCAGAUACAUUUGCUUGUGCGGUUGUAAAACAAGUAUUUUGUAUAAUUAUGCA ......-------.(.((((((((((.....)))))))...))).)(((.(((((..(((((((...........)))))))...))))).....))). ( -26.20) >DroSec_CAF1 16198 99 - 1 AGACUUUUGACUUUUGACAAGACAGGGCCAACCUGUCUACAUCUGAGCAGAUACAUUUGCGUGUGCGGUUGUAAAACAAGUAUUUUGUAUAAUUAUGCA .....((((.(((..((..(((((((.....)))))))...)).)))))))((((.((((....)))).))))......((((..(.....)..)))). ( -21.80) >DroSim_CAF1 16143 92 - 1 AGACUU-------UUGACAAGACAGGGCCAACCUGUCUACAUGUGAGCAGAUACAUUUGCGUGUGCGGUUGUAAAACAAGUAUUUUGUAUAAUUAUGCA ......-------.(.((((((((((.....)))))))...))).)(((.(((((..(((.(((...........))).)))...))))).....))). ( -22.10) >DroEre_CAF1 16606 91 - 1 AGACU--------UUGACAAGACAGGGCCAACCUGUCUACAUGAGAGCAGAUACAUUUGCGUGUGCGGUUGUAAAACAAGUAUAAUAUAUAAUUAUGCA .(.((--------((.(..(((((((.....)))))))...).)))))...((((.((((....)))).))))......(((((((.....))))))). ( -23.20) >DroYak_CAF1 16924 92 - 1 AGACUU-------UUGACAAGACAGGGCCAACCUGUCUACAAGUGAGCAGAUACAUUUGAGAGUGCGGUUGUAAAACAAUGAUUGUGUAUAAUUAUGCA ..((((-------(.....(((((((.....))))))).((((((........)))))))))))(((((..(......)..)))))((((....)))). ( -22.90) >DroAna_CAF1 17729 75 - 1 AGACUU-------UUGACAUGACAGGGACAACCUGUCUGCUUAUGACCGGGUAUAGAUAUG--UAC-------AAAUAAAAAUAAU--------GUGCA .....(-------(((((((((((((.....))))))(((((......))))).....)))--).)-------)))..........--------..... ( -15.30) >consensus AGACUU_______UUGACAAGACAGGGCCAACCUGUCUACAUGUGAGCAGAUACAUUUGCGUGUGCGGUUGUAAAACAAGUAUUUUGUAUAAUUAUGCA ...................(((((((.....)))))))........(((.(((((..(((.(((...........))).)))...))))).....))). (-13.54 = -14.90 + 1.36)


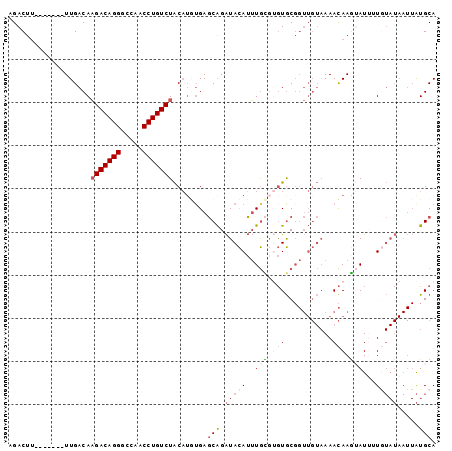
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:34:50 2006