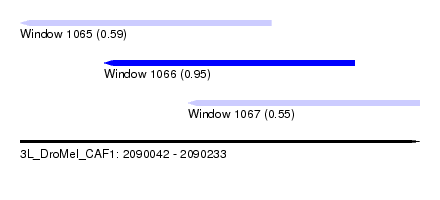
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 2,090,042 – 2,090,233 |
| Length | 191 |
| Max. P | 0.954318 |

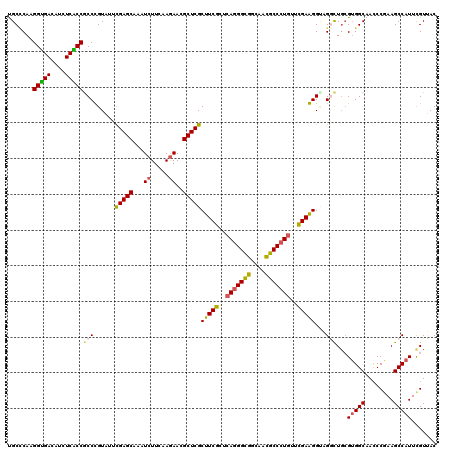
| Location | 2,090,042 – 2,090,162 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.44 |
| Mean single sequence MFE | -45.70 |
| Consensus MFE | -38.11 |
| Energy contribution | -37.67 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.594488 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2090042 120 - 23771897 UUCAAGAACGCUCGCUUCGCCCAGGGCGGCAAUGCCCUGUUCGAGGGACGCCUGCGUGGCAACCCGAAGCCAUUCGUGACCUGGACCCGAAAAGGCGCACCCCUUCUCGAGUCGCAGAAG .(((.(((.(...((((((..(((((((....)))))))..)..(((..(((.....)))..))))))))).))).))).((((((.(((.((((......)))).))).))).)))... ( -46.80) >DroVir_CAF1 167747 120 - 1 UUCAAGAACGCUCGCUUCGCUCAGGGCGGCAACGCCCUGUUUGAAGGUAGGCUGCGUGGCAACCCGAAGCCAUUCGUUACAUGGACCCGAAAAGGCGCACCACUUCUCGAGUCGCAAAAG ......((((....(((((..(((((((....)))))))..)))))...((((.((.(....).)).))))...))))...(((((.(((.(((........))).))).))).)).... ( -45.60) >DroGri_CAF1 142385 120 - 1 UUUAAGAACGCUCGCUUCGCUCAGGGUGGCAACGCCCUGUUCGAAGGUCGGCUGCGUGGCAACCCAAAGCCAUUCGUUACAUGGACCCGAAAAGGCGCACCACUUCUCGAGUCGCAAAAG ........(((((((((((..(((((((....)))))))..)))))...((.((((((((........))).((((...........))))...)))))))......)))).))...... ( -43.80) >DroWil_CAF1 186547 120 - 1 UUUAAAAACGCUCGCUUCGCUCAAGGCGGCAACGCCCUCUUUGAGGGUAGACUGCGUGGCAAUCCAAAGCCAUUUGUUACUUGGACCCGAAAAGGCGCACCACUUCUCGAGUCGCAAAAG .......((((...((.(.(((((((((....))))....))))).).))...))))(((........))).........((((((.(((.(((........))).))).))).)))... ( -37.20) >DroMoj_CAF1 146188 120 - 1 UUUAAGAACGCUCGCUUCGCUCAGGGUGGCAACGCCCUGUUCGAAGGCAGGCUCCGUGGCAACCCGAAGCCAUUCGUUACAUGGACCCGAAAAGGCGCACCACUUCUCGAGUCGCAAAAG ......((((...((((((..(((((((....)))))))..)))).)).((((.((.(....).)).))))...))))...(((((.(((.(((........))).))).))).)).... ( -45.30) >DroAna_CAF1 149056 120 - 1 UUCAAGAACGCUCGCUUCGCCCAGGGCGGCAACGCCCUGUUCGAGGGCCGGUUGAGGGGCAACCCGAAGCCAUUCGUGACAUGGACGCGGAAAGGCGCACCCCUUCUCGAGUCGCAGAAG ........(((((((((((..(((((((....)))))))..)))))(((((((..(((....)))..)))).((((((.......))))))..)))...........)))).))...... ( -55.50) >consensus UUCAAGAACGCUCGCUUCGCUCAGGGCGGCAACGCCCUGUUCGAAGGUAGGCUGCGUGGCAACCCGAAGCCAUUCGUUACAUGGACCCGAAAAGGCGCACCACUUCUCGAGUCGCAAAAG .........((...(((((..(((((((....)))))))..)))))...((((...(((....))).))))............(((.(((.(((........))).))).)))))..... (-38.11 = -37.67 + -0.44)

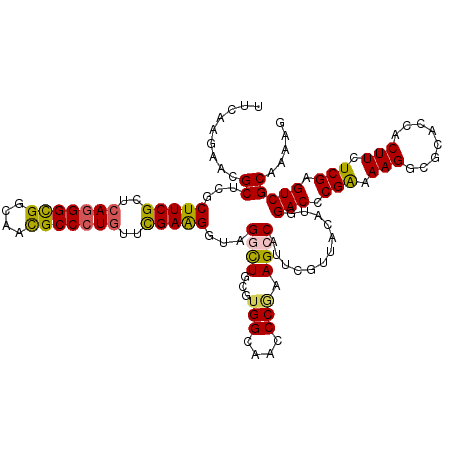
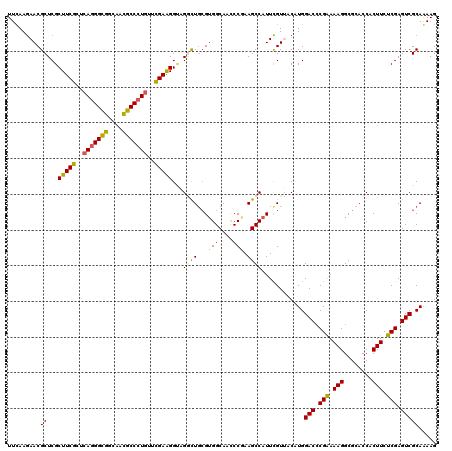
| Location | 2,090,082 – 2,090,202 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.89 |
| Mean single sequence MFE | -50.92 |
| Consensus MFE | -41.21 |
| Energy contribution | -40.63 |
| Covariance contribution | -0.58 |
| Combinations/Pair | 1.25 |
| Mean z-score | -3.40 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.44 |
| SVM RNA-class probability | 0.954318 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
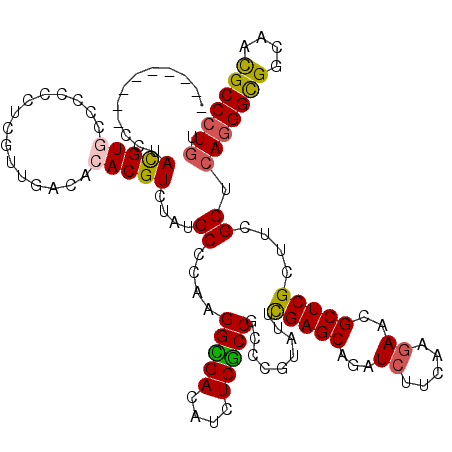
>3L_DroMel_CAF1 2090082 120 - 23771897 CGCCCAGGGCGACAUCUCGCCGCCCGUCUUCGAGCAGAUCUUCAAGAACGCUCGCUUCGCCCAGGGCGGCAAUGCCCUGUUCGAGGGACGCCUGCGUGGCAACCCGAAGCCAUUCGUGAC ((..((((((((....))(((((((.....(((((.(.((.....)).)))))).........)))))))...))))))..)).(((..(((.....)))..)))(((....)))..... ( -49.24) >DroVir_CAF1 167787 120 - 1 UGCGCAAGGUGACAUCUCACCGCCUGUAUUUGAGCAAAUCUUCAAGAACGCUCGCUUCGCUCAGGGCGGCAACGCCCUGUUUGAAGGUAGGCUGCGUGGCAACCCGAAGCCAUUCGUUAC .(((((.(((((....)))))(((((.....((((...((.....))..)))).(((((..(((((((....)))))))..))))).))))))))))(((........)))......... ( -52.90) >DroGri_CAF1 142425 120 - 1 UGCCCAAGGUGACAUUUCACCGCCUGUAUUCGAGCAGAUCUUUAAGAACGCUCGCUUCGCUCAGGGUGGCAACGCCCUGUUCGAAGGUCGGCUGCGUGGCAACCCAAAGCCAUUCGUUAC .((....(((((....)))))(((......(((((.(.((.....)).))))))(((((..(((((((....)))))))..)))))...))).))(((((........)))))....... ( -48.70) >DroWil_CAF1 186587 120 - 1 UGCUCAAGGUGAUAUUUCGCCACCCGUUUUCGAGCAAAUCUUUAAAAACGCUCGCUUCGCUCAAGGCGGCAACGCCCUCUUUGAGGGUAGACUGCGUGGCAAUCCAAAGCCAUUUGUUAC (((((..(((((....)))))....(....))))))...........((((...((.(.(((((((((....))))....))))).).))...))))(((........)))......... ( -39.70) >DroMoj_CAF1 146228 120 - 1 UGCCCAAGGUGACAUCUCACCGCCUGUAUUCGAGCAAAUCUUUAAGAACGCUCGCUUCGCUCAGGGUGGCAACGCCCUGUUCGAAGGCAGGCUCCGUGGCAACCCGAAGCCAUUCGUUAC (((((..(((((....)))))((((((...(((((...((.....))..)))))(((((..(((((((....)))))))..)))))))))))...).))))...((((....)))).... ( -54.80) >DroAna_CAF1 149096 120 - 1 CGCCCAGGGCGACAUCUCGCCACCCGUUUUUGAGCAAAUCUUCAAGAACGCUCGCUUCGCCCAGGGCGGCAACGCCCUGUUCGAGGGCCGGUUGAGGGGCAACCCGAAGCCAUUCGUGAC .((((..(((((....)))))...((((((((((......))))))))))......(((..(((((((....)))))))..))))))).((((..(((....)))..))))......... ( -60.20) >consensus UGCCCAAGGUGACAUCUCACCGCCCGUAUUCGAGCAAAUCUUCAAGAACGCUCGCUUCGCUCAGGGCGGCAACGCCCUGUUCGAAGGUAGGCUGCGUGGCAACCCGAAGCCAUUCGUUAC .......(((((....)))))(((......(((((...((.....))..)))))(((((..(((((((....)))))))..)))))...)))...(((((........)))))....... (-41.21 = -40.63 + -0.58)

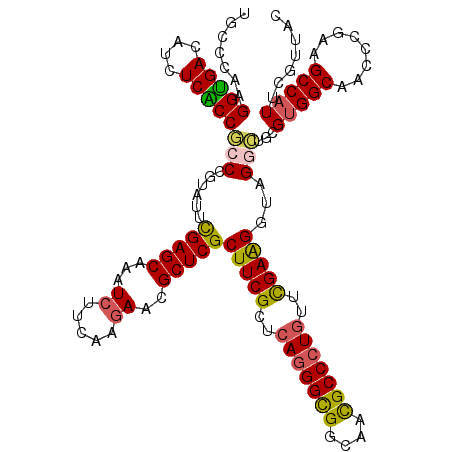
| Location | 2,090,122 – 2,090,233 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.25 |
| Mean single sequence MFE | -38.76 |
| Consensus MFE | -27.81 |
| Energy contribution | -27.39 |
| Covariance contribution | -0.41 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.547847 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
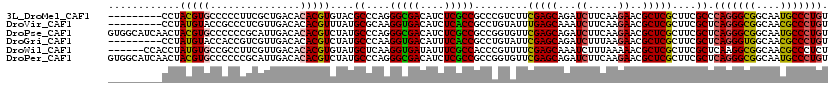
>3L_DroMel_CAF1 2090122 111 - 23771897 ---------CCUACGUGCCCCCUUCGCUGACACACGUGUACGCCCAGGGCGACAUCUCGCCGCCCGUCUUCGAGCAGAUCUUCAAGAACGCUCGCUUCGCCCAGGGCGGCAAUGCCCUGU ---------..((((((...............))))))......((((((((....))(((((((.....(((((.(.((.....)).)))))).........)))))))...)))))). ( -39.10) >DroVir_CAF1 167827 111 - 1 ---------CCUAUGUACCGCCCUCGUUGACACACGUUUAUGCGCAAGGUGACAUCUCACCGCCUGUAUUUGAGCAAAUCUUCAAGAACGCUCGCUUCGCUCAGGGCGGCAACGCCCUGU ---------..........((....((.(((....)))...))((..(((((....)))))..........((((...((.....))..))))))...)).(((((((....))))))). ( -33.10) >DroPse_CAF1 179532 120 - 1 GUGGCAUCAACUACGUGCCCCCCGCAUUGACACACGUCUAUGCCCAGGGCGACAUCUCGCCGCCGGUGUUCGAGCAGAUCUUCAAGAACGCUCGCUUCGCUCAGGGCGGCAAUGCCCUGU ..(((((.......)))))....((((.(((....))).)))).((((((((....))((((((((((..(((((.(.((.....)).))))))...))))...))))))...)))))). ( -48.30) >DroGri_CAF1 142465 111 - 1 ---------CCUAUGUACCACCGUCGUUGACACACGUCUAUGCCCAAGGUGACAUUUCACCGCCUGUAUUCGAGCAGAUCUUUAAGAACGCUCGCUUCGCUCAGGGUGGCAACGCCCUGU ---------.....((((....(.(((.(((....))).))))....(((((....)))))....)))).(((((.(.((.....)).)))))).......(((((((....))))))). ( -31.80) >DroWil_CAF1 186627 114 - 1 ------CCACCUAUGUGCCGCCUUCGUUGACACACGUGUAUGCUCAAGGUGAUAUUUCGCCACCCGUUUUCGAGCAAAUCUUUAAAAACGCUCGCUUCGCUCAAGGCGGCAACGCCCUCU ------.........(((((((((.((......))(((...((....(((((....)))))..........((((..............))))))..)))..)))))))))......... ( -31.94) >DroPer_CAF1 157328 120 - 1 GUGGCAUCAACUACGUGCCCCCCGCAUUGACACACGUCUAUGCCCAGGGCGACAUCUCGCCGCCGGUGUUCGAGCAGAUCUUCAAGAACGCUCGCUUCGCUCAGGGCGGCAAUGCCCUGU ..(((((.......)))))....((((.(((....))).)))).((((((((....))((((((((((..(((((.(.((.....)).))))))...))))...))))))...)))))). ( -48.30) >consensus _________CCUACGUGCCCCCCUCGUUGACACACGUCUAUGCCCAAGGCGACAUCUCGCCGCCCGUAUUCGAGCAGAUCUUCAAGAACGCUCGCUUCGCUCAGGGCGGCAACGCCCUGU ............(((((...............)))))....((....(((((....))))).........(((((...((.....))..)))))....)).(((((((....))))))). (-27.81 = -27.39 + -0.41)
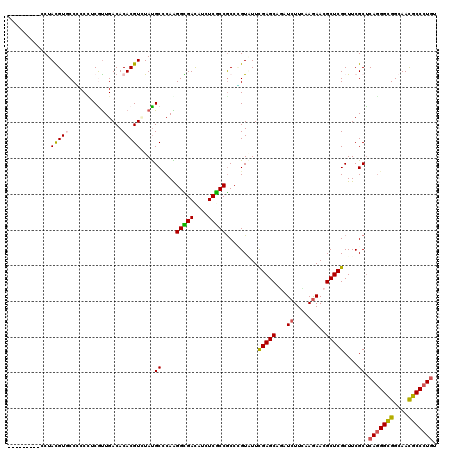
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:48:49 2006