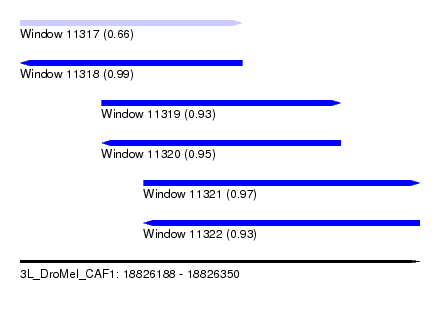
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 18,826,188 – 18,826,350 |
| Length | 162 |
| Max. P | 0.986410 |

| Location | 18,826,188 – 18,826,278 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 87.50 |
| Mean single sequence MFE | -19.25 |
| Consensus MFE | -17.40 |
| Energy contribution | -17.40 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.656809 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18826188 90 + 23771897 UGGCUA----AGUUUGGCUGUUUUAUAUAAGAGUUCCUUCACGGACACUUAUAUAAUUUUGAUUGUUGCUCAGAAAGCCCUCAUUGUUUACCAG (((...----(((..((((...(((((((((.((((......)))).)))))))))((((((.......))))))))))...))).....))). ( -19.20) >DroEre_CAF1 12197 89 + 1 UGGCUA----AGUUUGGUUGUUUUAUAUAAGAGUUCCUACACGGACACUUAUAUAAUUUUGAUUGUUGCUCAGAAAGCCC-UAUUAUCUAACAG .((((.----............(((((((((.((((......)))).)))))))))((((((.......)))))))))).-............. ( -18.20) >DroYak_CAF1 1365 89 + 1 UGGCUA----AGUUUGGUUUUUUUAUAUAAGAGUUCC-GCACGGACACUUAUAUAAUUUUGAUUGUUGCUCAGAAAGCCCUUACUAUUUAACAG .((((.----............(((((((((.((.((-....)))).)))))))))((((((.......))))))))))............... ( -18.20) >DroAna_CAF1 1700 94 + 1 UGGCUGUAAUAGUUUGGCUUUUUUAUAUAAGAGCUCCUAAAUGGACACUUAUAUAAUUUUGAUUGUUGCUCAGAAAGCCCUUAUAAAUUAUUAG ......(((((((((((((((((((((((((.(.(((.....)))).)))))))))...(((.......)))))))))).....))))))))). ( -21.40) >consensus UGGCUA____AGUUUGGCUGUUUUAUAUAAGAGUUCCUACACGGACACUUAUAUAAUUUUGAUUGUUGCUCAGAAAGCCCUUAUUAUUUAACAG .((((.................(((((((((.(.(((.....)))).)))))))))((((((.......))))))))))............... (-17.40 = -17.40 + 0.00)


| Location | 18,826,188 – 18,826,278 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 87.50 |
| Mean single sequence MFE | -18.68 |
| Consensus MFE | -16.96 |
| Energy contribution | -16.77 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.91 |
| SVM decision value | 2.04 |
| SVM RNA-class probability | 0.986410 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18826188 90 - 23771897 CUGGUAAACAAUGAGGGCUUUCUGAGCAACAAUCAAAAUUAUAUAAGUGUCCGUGAAGGAACUCUUAUAUAAAACAGCCAAACU----UAGCCA .((((......(((..((((...)))).....)))...(((((((((..(((.....)))...)))))))))....))))....----...... ( -19.40) >DroEre_CAF1 12197 89 - 1 CUGUUAGAUAAUA-GGGCUUUCUGAGCAACAAUCAAAAUUAUAUAAGUGUCCGUGUAGGAACUCUUAUAUAAAACAACCAAACU----UAGCCA (((((....))))-)((((...(((.......)))...(((((((((..(((.....)))...)))))))))............----.)))). ( -19.10) >DroYak_CAF1 1365 89 - 1 CUGUUAAAUAGUAAGGGCUUUCUGAGCAACAAUCAAAAUUAUAUAAGUGUCCGUGC-GGAACUCUUAUAUAAAAAAACCAAACU----UAGCCA ..((((...(((..((((((...))))...........(((((((((..(((....-)))...))))))))).....))..)))----)))).. ( -17.60) >DroAna_CAF1 1700 94 - 1 CUAAUAAUUUAUAAGGGCUUUCUGAGCAACAAUCAAAAUUAUAUAAGUGUCCAUUUAGGAGCUCUUAUAUAAAAAAGCCAAACUAUUACAGCCA ...............((((((.(((.......)))...(((((((((.((((.....))).).))))))))).))))))............... ( -18.60) >consensus CUGUUAAAUAAUAAGGGCUUUCUGAGCAACAAUCAAAAUUAUAUAAGUGUCCGUGUAGGAACUCUUAUAUAAAAAAACCAAACU____UAGCCA ...............((((...(((.......)))...(((((((((.((((.....)).)).))))))))).................)))). (-16.96 = -16.77 + -0.19)



| Location | 18,826,221 – 18,826,318 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 80.40 |
| Mean single sequence MFE | -37.29 |
| Consensus MFE | -37.31 |
| Energy contribution | -36.73 |
| Covariance contribution | -0.58 |
| Combinations/Pair | 1.16 |
| Mean z-score | -7.01 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.932546 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18826221 97 + 23771897 U-------------UCACGGACACUUAUAUAAUUUUGAUUGUUGCUCAGAAAGCCCUCAUUGUUUACCAGUUGGCUUUCGAGCAAUAAUUGAAACCAGAUAAGUGAUCCC .-------------....((((((((((....(((..((((((((((.(((((((...((((.....)))).)))))))))))))))))..)))....))))))).))). ( -40.00) >DroVir_CAF1 10597 102 + 1 ------ACCUCACAAAACGGACGCCUAUAUCAUUUUGAUUGUUGCUCAGAAAGCCCUG--CAUUCAAUACUUGGCUUUCGAGUAAUAAUCGAAACCAAAUAAGCGAUCCU ------............((((((.(((....(((((((((((((((.(((((((..(--.........)..))))))))))))))))))))))....))).))).))). ( -35.00) >DroEre_CAF1 12230 96 + 1 U-------------ACACGGACACUUAUAUAAUUUUGAUUGUUGCUCAGAAAGCCC-UAUUAUCUAACAGCUGGCUUUCGAGCAAUAAUUGAAACCAGAUAAGUGAUCCU .-------------....((((((((((....(((..((((((((((.(((((((.-...............)))))))))))))))))..)))....))))))).))). ( -38.09) >DroYak_CAF1 1398 96 + 1 --------------GCACGGACACUUAUAUAAUUUUGAUUGUUGCUCAGAAAGCCCUUACUAUUUAACAGCUGGCUUUCGAGCAAUAAUUGAAACCAGAUAAGUGAUCCU --------------....((((((((((....(((..((((((((((.(((((((.................)))))))))))))))))..)))....))))))).))). ( -38.03) >DroAna_CAF1 1737 97 + 1 U-------------AAAUGGACACUUAUAUAAUUUUGAUUGUUGCUCAGAAAGCCCUUAUAAAUUAUUAGCUGGCUUUCGAGCAAUAAUUGAAACCAAAUAAGUGAUCCU .-------------....((((((((((....(((..((((((((((.(((((((....(((....)))...)))))))))))))))))..)))....))))))).))). ( -37.50) >DroPer_CAF1 19752 110 + 1 AUUGUGAAAGUAUAAACCGGACACUUAUCUAUUUUUGAUUGUUGCUCAGAAAGCCCUUAUAAUUAACCAGUUGGCUUUCGAGCAAUUAUCAAAGCCAAAUAAGUGAUCCC ..................((((((((((....(((((((.(((((((.(((((((.................)))))))))))))).)))))))....))))))).))). ( -35.13) >consensus U_____________AAACGGACACUUAUAUAAUUUUGAUUGUUGCUCAGAAAGCCCUUAUUAUUUAACAGCUGGCUUUCGAGCAAUAAUUGAAACCAAAUAAGUGAUCCU ..................((((((((((....(((((((((((((((.(((((((.................))))))))))))))))))))))....))))))).))). (-37.31 = -36.73 + -0.58)


| Location | 18,826,221 – 18,826,318 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 80.40 |
| Mean single sequence MFE | -36.60 |
| Consensus MFE | -31.90 |
| Energy contribution | -32.31 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.06 |
| Mean z-score | -5.83 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.953046 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18826221 97 - 23771897 GGGAUCACUUAUCUGGUUUCAAUUAUUGCUCGAAAGCCAACUGGUAAACAAUGAGGGCUUUCUGAGCAACAAUCAAAAUUAUAUAAGUGUCCGUGA-------------A .(((.(((((((.((((((..(((.(((((((((((((.(.((.....)).)...))))))).)))))).)))..)))))).))))))))))....-------------. ( -35.40) >DroVir_CAF1 10597 102 - 1 AGGAUCGCUUAUUUGGUUUCGAUUAUUACUCGAAAGCCAAGUAUUGAAUG--CAGGGCUUUCUGAGCAACAAUCAAAAUGAUAUAGGCGUCCGUUUUGUGAGGU------ .(((.(((((((.(.((((.((((.((.((((((((((..((((...)))--)..))))))).))).)).)))).)))).).))))))))))............------ ( -34.60) >DroEre_CAF1 12230 96 - 1 AGGAUCACUUAUCUGGUUUCAAUUAUUGCUCGAAAGCCAGCUGUUAGAUAAUA-GGGCUUUCUGAGCAACAAUCAAAAUUAUAUAAGUGUCCGUGU-------------A .(((.(((((((.((((((..(((.(((((((((((((..(((((....))))-)))))))).)))))).)))..)))))).))))))))))....-------------. ( -37.10) >DroYak_CAF1 1398 96 - 1 AGGAUCACUUAUCUGGUUUCAAUUAUUGCUCGAAAGCCAGCUGUUAAAUAGUAAGGGCUUUCUGAGCAACAAUCAAAAUUAUAUAAGUGUCCGUGC-------------- .(((.(((((((.((((((..(((.(((((((((((((.(((((...)))))...))))))).)))))).)))..)))))).))))))))))....-------------- ( -38.10) >DroAna_CAF1 1737 97 - 1 AGGAUCACUUAUUUGGUUUCAAUUAUUGCUCGAAAGCCAGCUAAUAAUUUAUAAGGGCUUUCUGAGCAACAAUCAAAAUUAUAUAAGUGUCCAUUU-------------A .(((.(((((((.((((((..(((.(((((((((((((...(((....)))....))))))).)))))).)))..)))))).))))))))))....-------------. ( -35.20) >DroPer_CAF1 19752 110 - 1 GGGAUCACUUAUUUGGCUUUGAUAAUUGCUCGAAAGCCAACUGGUUAAUUAUAAGGGCUUUCUGAGCAACAAUCAAAAAUAGAUAAGUGUCCGGUUUAUACUUUCACAAU .(((.((((((((((..((((((..(((((((((((((.....((.....))...))))))).))))))..))))))..))))))))))))).................. ( -39.20) >consensus AGGAUCACUUAUCUGGUUUCAAUUAUUGCUCGAAAGCCAACUGUUAAAUAAUAAGGGCUUUCUGAGCAACAAUCAAAAUUAUAUAAGUGUCCGUGU_____________A .(((.(((((((.((((((..(((.(((((((((((((.................))))))).)))))).)))..)))))).)))))))))).................. (-31.90 = -32.31 + 0.42)



| Location | 18,826,238 – 18,826,350 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 79.93 |
| Mean single sequence MFE | -29.51 |
| Consensus MFE | -25.63 |
| Energy contribution | -25.30 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.14 |
| Mean z-score | -3.23 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.70 |
| SVM RNA-class probability | 0.972927 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18826238 112 + 23771897 AAUUUUGAUUGUUGCUCAGAAAGCCCUCAUUGUUUACCAGUUGGCUUUCGAGCAAUAAUUGAAACCAGAUAAGUGAUCCCCUCAACGCUGACGAUUUCAAAGCACUUCGUUU--- ..(((..((((((((((.(((((((...((((.....)))).)))))))))))))))))..))).(((.....(((.....)))...)))((((............))))..--- ( -32.40) >DroPse_CAF1 19354 112 + 1 AUUUUUGAUUGUUGCUCAGAAAGCCCUUAUAAUUAACCAGUUGGCUUUCGAGCAAUUAUCAAAGCCAAAUAAGUGAUCCCAACAACAUUUUAG-UGUGAAAU--ACUUGACUCAA ..(((((((.(((((((.(((((((.................)))))))))))))).))))))).....((((((.........((((....)-)))....)--)))))...... ( -27.05) >DroEre_CAF1 12247 111 + 1 AAUUUUGAUUGUUGCUCAGAAAGCCC-UAUUAUCUAACAGCUGGCUUUCGAGCAAUAAUUGAAACCAGAUAAGUGAUCCUUACAACGCUGAAGAUUUGACUGCAUUUCGUGU--- ..(((..((((((((((.(((((((.-...............)))))))))))))))))..))).(((((.((((..........))))....)))))..............--- ( -30.29) >DroYak_CAF1 1414 112 + 1 AAUUUUGAUUGUUGCUCAGAAAGCCCUUACUAUUUAACAGCUGGCUUUCGAGCAAUAAUUGAAACCAGAUAAGUGAUCCUUACAACAUUGAAGAUUUGACUGCGUUUCGUGU--- ......(((((((((((.(((((((.................))))))))))))))))))((((((((.(((((..((...........))..))))).))).)))))....--- ( -31.43) >DroAna_CAF1 1754 113 + 1 AAUUUUGAUUGUUGCUCAGAAAGCCCUUAUAAAUUAUUAGCUGGCUUUCGAGCAAUAAUUGAAACCAAAUAAGUGAUCCUGACAACAUUUUAAGUGUGAAAU--AUUCUUUUAAA ..(((..((((((((((.(((((((....(((....)))...)))))))))))))))))..))).....((((.((........((((.....)))).....--..)).)))).. ( -28.86) >DroPer_CAF1 19782 112 + 1 AUUUUUGAUUGUUGCUCAGAAAGCCCUUAUAAUUAACCAGUUGGCUUUCGAGCAAUUAUCAAAGCCAAAUAAGUGAUCCCAACAACAUUUUAG-UGUGAAAU--ACUUGACUCAA ..(((((((.(((((((.(((((((.................)))))))))))))).))))))).....((((((.........((((....)-)))....)--)))))...... ( -27.05) >consensus AAUUUUGAUUGUUGCUCAGAAAGCCCUUAUAAUUUACCAGCUGGCUUUCGAGCAAUAAUUGAAACCAAAUAAGUGAUCCCAACAACAUUGAAGAUGUGAAAG__AUUCGUCU___ ..(((((((((((((((.(((((((.................))))))))))))))))))))))................................................... (-25.63 = -25.30 + -0.33)


| Location | 18,826,238 – 18,826,350 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 79.93 |
| Mean single sequence MFE | -27.89 |
| Consensus MFE | -18.28 |
| Energy contribution | -18.33 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.66 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.926921 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18826238 112 - 23771897 ---AAACGAAGUGCUUUGAAAUCGUCAGCGUUGAGGGGAUCACUUAUCUGGUUUCAAUUAUUGCUCGAAAGCCAACUGGUAAACAAUGAGGGCUUUCUGAGCAACAAUCAAAAUU ---.........(((.((....))..))).((((.(..((((......))))..).....(((((((((((((.(.((.....)).)...))))))).))))))...)))).... ( -27.60) >DroPse_CAF1 19354 112 - 1 UUGAGUCAAGU--AUUUCACA-CUAAAAUGUUGUUGGGAUCACUUAUUUGGCUUUGAUAAUUGCUCGAAAGCCAACUGGUUAAUUAUAAGGGCUUUCUGAGCAACAAUCAAAAAU ..(((((((((--(.......-((((.......)))).......))))))))))((((..(((((((((((((.....((.....))...))))))).))))))..))))..... ( -31.04) >DroEre_CAF1 12247 111 - 1 ---ACACGAAAUGCAGUCAAAUCUUCAGCGUUGUAAGGAUCACUUAUCUGGUUUCAAUUAUUGCUCGAAAGCCAGCUGUUAGAUAAUA-GGGCUUUCUGAGCAACAAUCAAAAUU ---..(((...((.((......)).)).))).(((((.....)))))..(((((..(((.(((((((((((((..(((((....))))-)))))))).)))))).)))..))))) ( -26.60) >DroYak_CAF1 1414 112 - 1 ---ACACGAAACGCAGUCAAAUCUUCAAUGUUGUAAGGAUCACUUAUCUGGUUUCAAUUAUUGCUCGAAAGCCAGCUGUUAAAUAGUAAGGGCUUUCUGAGCAACAAUCAAAAUU ---....(((((.(((................(((((.....))))))))))))).....(((((((((((((.(((((...)))))...))))))).))))))........... ( -29.99) >DroAna_CAF1 1754 113 - 1 UUUAAAAGAAU--AUUUCACACUUAAAAUGUUGUCAGGAUCACUUAUUUGGUUUCAAUUAUUGCUCGAAAGCCAGCUAAUAAUUUAUAAGGGCUUUCUGAGCAACAAUCAAAAUU ........(((--((((........)))))))....((((((......))))))......(((((((((((((...(((....)))....))))))).))))))........... ( -21.10) >DroPer_CAF1 19782 112 - 1 UUGAGUCAAGU--AUUUCACA-CUAAAAUGUUGUUGGGAUCACUUAUUUGGCUUUGAUAAUUGCUCGAAAGCCAACUGGUUAAUUAUAAGGGCUUUCUGAGCAACAAUCAAAAAU ..(((((((((--(.......-((((.......)))).......))))))))))((((..(((((((((((((.....((.....))...))))))).))))))..))))..... ( -31.04) >consensus ___AAACGAAU__AUUUCAAAUCUAAAAUGUUGUAAGGAUCACUUAUCUGGUUUCAAUUAUUGCUCGAAAGCCAACUGGUAAAUAAUAAGGGCUUUCUGAGCAACAAUCAAAAUU .............................((((...((((((......))))))......(((((((((((((...((........))..))))))).))))))))))....... (-18.28 = -18.33 + 0.06)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:34:41 2006