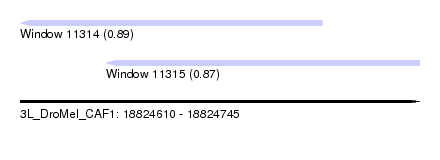
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 18,824,610 – 18,824,745 |
| Length | 135 |
| Max. P | 0.892645 |

| Location | 18,824,610 – 18,824,712 |
|---|---|
| Length | 102 |
| Sequences | 3 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 97.41 |
| Mean single sequence MFE | -35.80 |
| Consensus MFE | -33.50 |
| Energy contribution | -33.83 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.892645 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
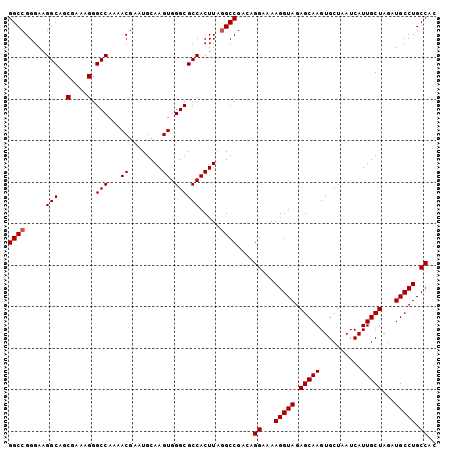
>3L_DroMel_CAF1 18824610 102 - 23771897 GGCCGGGAAGGCAGCGAAAGGGCCA-AACGAAUGCAAGUGGGCGCCACUUAAGCCGAAAGGAAAAGGUAGAGCAAGUGCUAAUCAUUGCUAGAUGCCUGCCAC (((((....(((..(....).))).-..)).....((((((...))))))..)))....((...(((((.(((((..........)))))...))))).)).. ( -32.00) >DroSec_CAF1 31996 103 - 1 GGCCGGGAAGGCAGCGAAAGGGCCAAAACGAAUGCAAGUGGGCGCCACUUCGGCCGACAGGAAAAGGUAGAGCAAGUGCUAAUCAUUGCUAGAUGCCUGCCAC (((((((..(((..(....).(((...((........)).))))))..)))))))....((...(((((.(((((..........)))))...))))).)).. ( -39.50) >DroSim_CAF1 33133 103 - 1 GGCCGGGAAGGCAGCGAAAGGGCCAAAACGAAUGCAAGUGGGCGCCACUUAGGCCGACAGGAAAAGGUAGAGCAAGUGCUAAUCAUUGCUAGAUGCCUGCCAC ((((.....(((..(....).)))...........((((((...)))))).))))....((...(((((.(((((..........)))))...))))).)).. ( -35.90) >consensus GGCCGGGAAGGCAGCGAAAGGGCCAAAACGAAUGCAAGUGGGCGCCACUUAGGCCGACAGGAAAAGGUAGAGCAAGUGCUAAUCAUUGCUAGAUGCCUGCCAC ((((.....(((..(....).(((...((........)).)))))).....))))....((...(((((.(((((..........)))))...))))).)).. (-33.50 = -33.83 + 0.33)


| Location | 18,824,639 – 18,824,745 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 97.51 |
| Mean single sequence MFE | -33.27 |
| Consensus MFE | -30.90 |
| Energy contribution | -31.23 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.867631 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
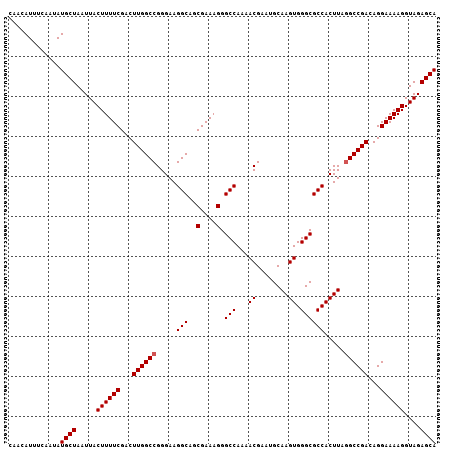
>3L_DroMel_CAF1 18824639 106 - 23771897 CAACAUUUCAAUAUGCUAAUUACUUUUCGACUUGGCCGGGAAGGCAGCGAAAGGGCCA-AACGAAUGCAAGUGGGCGCCACUUAAGCCGAAAGGAAAAGGUAGAGCA .............((((.....(((((((.....(((.....)))..)))))))(((.-.........((((((...))))))...((....))....)))..)))) ( -29.60) >DroSec_CAF1 32025 107 - 1 CAACAUUUCAAUAUGCUAAUUACUUUUCGACUUGGCCGGGAAGGCAGCGAAAGGGCCAAAACGAAUGCAAGUGGGCGCCACUUCGGCCGACAGGAAAAGGUAGAGCA .............((((.....((((((...(((((((((..(((..(....).(((...((........)).))))))..)))))))))...))))))....)))) ( -36.90) >DroSim_CAF1 33162 107 - 1 CAACAUUUCAAUAUGCUAAUUACUUUUCGACUUGGCCGGGAAGGCAGCGAAAGGGCCAAAACGAAUGCAAGUGGGCGCCACUUAGGCCGACAGGAAAAGGUAGAGCA .............((((.....((((((...((((((.....(((..(....).)))...........((((((...)))))).))))))...))))))....)))) ( -33.30) >consensus CAACAUUUCAAUAUGCUAAUUACUUUUCGACUUGGCCGGGAAGGCAGCGAAAGGGCCAAAACGAAUGCAAGUGGGCGCCACUUAGGCCGACAGGAAAAGGUAGAGCA .............((((.....((((((...((((((.....(((..(....).(((...((........)).)))))).....))))))...))))))....)))) (-30.90 = -31.23 + 0.33)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:34:35 2006