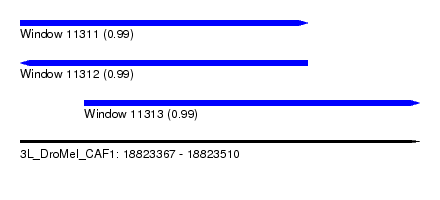
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 18,823,367 – 18,823,510 |
| Length | 143 |
| Max. P | 0.991518 |

| Location | 18,823,367 – 18,823,470 |
|---|---|
| Length | 103 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.06 |
| Mean single sequence MFE | -33.60 |
| Consensus MFE | -25.91 |
| Energy contribution | -27.47 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.06 |
| Mean z-score | -4.73 |
| Structure conservation index | 0.77 |
| SVM decision value | 2.15 |
| SVM RNA-class probability | 0.989058 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18823367 103 + 23771897 U-----------------AUAAAAACUAUAUUUAAUUAGCCUCGUAUUUGACACUGAUGUGCAUCAAAGCGAUUCCUUUAAUUUAGGGACGUUUUGAGCAAAGUCUUGAAUAAGAGGCUA .-----------------..................(((((((.((((..(.(((....(((.((((((((.(((((.......)))))))))))))))).))).)..)))).))))))) ( -36.60) >DroSec_CAF1 30657 120 + 1 UUCCUUUUUUGUUAUAGAAUAAAAACUAUAUUUAAUUAGCCUCUUAUUUGACACUGAUGUGCAUCAAAGCGAUUCCUUUAACUUAGGAACAUUAUGACCACUGUCUUGAAUAAGAGGCUG .....((((((((....))))))))...........((((((((((((..(...((((((((......))...((((.......)))))))))).(((....))))..)))))))))))) ( -30.00) >DroSim_CAF1 31755 120 + 1 UUCCUUUUUUGUUAGAGAAUAAAAACUAUAUUUAAUUAGCCUCUUAUUUGACACUGAUGUACAUCAAAGCGAUUCCUUUUAUUUAAGGACGUAAUGACCACAGUCUUGAAUAAGAGGCUA .....((((((((....))))))))...........((((((((((((..(.((((.((..(((....(((..(((((......)))))))).)))..)))))).)..)))))))))))) ( -34.20) >consensus UUCCUUUUUUGUUA_AGAAUAAAAACUAUAUUUAAUUAGCCUCUUAUUUGACACUGAUGUGCAUCAAAGCGAUUCCUUUAAUUUAGGGACGUUAUGACCACAGUCUUGAAUAAGAGGCUA ....................................((((((((((((..(.((((.((....(((.((((.(((((.......))))))))).))).)))))).)..)))))))))))) (-25.91 = -27.47 + 1.56)



| Location | 18,823,367 – 18,823,470 |
|---|---|
| Length | 103 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.06 |
| Mean single sequence MFE | -29.63 |
| Consensus MFE | -19.71 |
| Energy contribution | -20.60 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.79 |
| Structure conservation index | 0.67 |
| SVM decision value | 2.09 |
| SVM RNA-class probability | 0.987754 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18823367 103 - 23771897 UAGCCUCUUAUUCAAGACUUUGCUCAAAACGUCCCUAAAUUAAAGGAAUCGCUUUGAUGCACAUCAGUGUCAAAUACGAGGCUAAUUAAAUAUAGUUUUUAU-----------------A (((((((.((((..(.(((.((((((((.((..(((.......)))...)).))))).)))....))).)..)))).)))))))..................-----------------. ( -25.00) >DroSec_CAF1 30657 120 - 1 CAGCCUCUUAUUCAAGACAGUGGUCAUAAUGUUCCUAAGUUAAAGGAAUCGCUUUGAUGCACAUCAGUGUCAAAUAAGAGGCUAAUUAAAUAUAGUUUUUAUUCUAUAACAAAAAAGGAA .(((((((((((...((((.((((......((((((.......)))))).((......))..)))).)))).))))))))))).......(((((........)))))............ ( -32.90) >DroSim_CAF1 31755 120 - 1 UAGCCUCUUAUUCAAGACUGUGGUCAUUACGUCCUUAAAUAAAAGGAAUCGCUUUGAUGUACAUCAGUGUCAAAUAAGAGGCUAAUUAAAUAUAGUUUUUAUUCUCUAACAAAAAAGGAA ((((((((((((..(.((((((..((((((((((((......)))))..))...)))))..)).)))).)..)))))))))))).................................... ( -31.00) >consensus UAGCCUCUUAUUCAAGACUGUGGUCAUAACGUCCCUAAAUUAAAGGAAUCGCUUUGAUGCACAUCAGUGUCAAAUAAGAGGCUAAUUAAAUAUAGUUUUUAUUCU_UAACAAAAAAGGAA ((((((((((((..(.((((((...........(((.......)))....((......)).)).)))).)..)))))))))))).................................... (-19.71 = -20.60 + 0.89)


| Location | 18,823,390 – 18,823,510 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.98 |
| Mean single sequence MFE | -29.03 |
| Consensus MFE | -21.74 |
| Energy contribution | -23.63 |
| Covariance contribution | 1.89 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.09 |
| Structure conservation index | 0.75 |
| SVM decision value | 2.27 |
| SVM RNA-class probability | 0.991518 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18823390 120 + 23771897 CUCGUAUUUGACACUGAUGUGCAUCAAAGCGAUUCCUUUAAUUUAGGGACGUUUUGAGCAAAGUCUUGAAUAAGAGGCUAAUUAAAGCUGUGCCUAAUUAAAGUGUAUAUUCAACACUAA (((.((((..(.(((....(((.((((((((.(((((.......)))))))))))))))).))).)..)))).)))(((......))).............(((((.......))))).. ( -33.60) >DroSec_CAF1 30697 104 + 1 CUCUUAUUUGACACUGAUGUGCAUCAAAGCGAUUCCUUUAACUUAGGAACAUUAUGACCACUGUCUUGAAUAAGAGGCUGAU----------------UAAAGUGUAUAUUCUGCACUAA ((((((((..(...((((((((......))...((((.......)))))))))).(((....))))..))))))))......----------------...((((((.....)))))).. ( -24.80) >DroSim_CAF1 31795 104 + 1 CUCUUAUUUGACACUGAUGUACAUCAAAGCGAUUCCUUUUAUUUAAGGACGUAAUGACCACAGUCUUGAAUAAGAGGCUAAU----------------UAAAGUGUAUAUUCUGCACUAA ((((((((..(.((((.((..(((....(((..(((((......)))))))).)))..)))))).)..))))))))......----------------...((((((.....)))))).. ( -28.70) >consensus CUCUUAUUUGACACUGAUGUGCAUCAAAGCGAUUCCUUUAAUUUAGGGACGUUAUGACCACAGUCUUGAAUAAGAGGCUAAU________________UAAAGUGUAUAUUCUGCACUAA ((((((((..(.((((.((....(((.((((.(((((.......))))))))).))).)))))).)..)))))))).........................((((((.....)))))).. (-21.74 = -23.63 + 1.89)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:34:33 2006