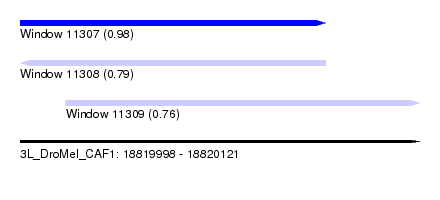
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 18,819,998 – 18,820,121 |
| Length | 123 |
| Max. P | 0.978499 |

| Location | 18,819,998 – 18,820,092 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 82.44 |
| Mean single sequence MFE | -28.65 |
| Consensus MFE | -22.86 |
| Energy contribution | -22.80 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.82 |
| SVM RNA-class probability | 0.978499 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
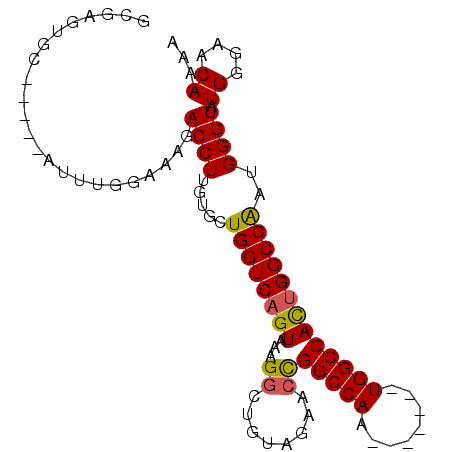
>3L_DroMel_CAF1 18819998 94 + 23771897 GCGAGUGC-----AUUUGGAAAGACCUUGUGCUGGUCAGUAAAGAAUGUAGAACCGGCCAAUGGCCAAUGGCCACUGGCCAAUGGGUAUGGCAACAAAA ((...(((-----((((..(..((((.......))))..)...)))))))..((((((((.(((((...))))).)))))....)))...))....... ( -32.50) >DroSec_CAF1 27521 87 + 1 GCGAGUGG-----AUUUGGAAAGACCUUGUGCUGGUCAGUAAAGGCUGUAGAACCGGCCAA-------UGGCCACUGGCCAAGGGGUAUGGGAACAAAA ((.(((((-----.(((....)))(((((.((((((((((....))))....)))))))))-------.))))))).)).........((....))... ( -27.10) >DroSim_CAF1 28151 87 + 1 GCGAGUGC-----AUUUGGAAAGACCUUGUGCUGGUCAGUAAAGGCUGUAGAACCGGCCAA-------UGGCCACUGGCCAACGGGUAUGGGAACAAAA .((...((-----((..((.....))..))))((((((((...((........))((((..-------.)))))))))))).))....((....))... ( -27.60) >DroAna_CAF1 19819 87 + 1 GCAAGAGGGCUAAAAAUGGAAAGACCUUGUGCUGGUCAGUAAAGGU----AAACUGGCCAA-------UGGCCAUCGGCCG-UGGGUAUGGGAACAAAA (((.((((.(((....))).....)))).)))((((((((......----..))))))))(-------(((((...)))))-).....((....))... ( -27.40) >consensus GCGAGUGC_____AUUUGGAAAGACCUUGUGCUGGUCAGUAAAGGCUGUAGAACCGGCCAA_______UGGCCACUGGCCAAUGGGUAUGGGAACAAAA .......................((((.....((((((((...((........))(((((........)))))))))))))..)))).((....))... (-22.86 = -22.80 + -0.06)


| Location | 18,819,998 – 18,820,092 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 82.44 |
| Mean single sequence MFE | -22.50 |
| Consensus MFE | -14.96 |
| Energy contribution | -15.28 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.793499 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18819998 94 - 23771897 UUUUGUUGCCAUACCCAUUGGCCAGUGGCCAUUGGCCAUUGGCCGGUUCUACAUUCUUUACUGACCAGCACAAGGUCUUUCCAAAU-----GCACUCGC ......(((.......((((((((((((((...))))))))))))))...............((((.......)))).........-----)))..... ( -30.30) >DroSec_CAF1 27521 87 - 1 UUUUGUUCCCAUACCCCUUGGCCAGUGGCCA-------UUGGCCGGUUCUACAGCCUUUACUGACCAGCACAAGGUCUUUCCAAAU-----CCACUCGC ...............(((((((((((((((.-------..))))(((......)))...))))....)).)))))...........-----........ ( -19.30) >DroSim_CAF1 28151 87 - 1 UUUUGUUCCCAUACCCGUUGGCCAGUGGCCA-------UUGGCCGGUUCUACAGCCUUUACUGACCAGCACAAGGUCUUUCCAAAU-----GCACUCGC ................(((((.((((((((.-------..))))(((......)))...)))).)))))....((.....))....-----........ ( -21.90) >DroAna_CAF1 19819 87 - 1 UUUUGUUCCCAUACCCA-CGGCCGAUGGCCA-------UUGGCCAGUUU----ACCUUUACUGACCAGCACAAGGUCUUUCCAUUUUUAGCCCUCUUGC .................-.((((...)))).-------.(((.((((..----......)))).)))(((..(((.((..........)).)))..))) ( -18.50) >consensus UUUUGUUCCCAUACCCAUUGGCCAGUGGCCA_______UUGGCCGGUUCUACAGCCUUUACUGACCAGCACAAGGUCUUUCCAAAU_____CCACUCGC .................((((....((((((........)))))).................((((.......))))...))))............... (-14.96 = -15.28 + 0.31)



| Location | 18,820,012 – 18,820,121 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.18 |
| Mean single sequence MFE | -30.48 |
| Consensus MFE | -23.98 |
| Energy contribution | -24.60 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.759958 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18820012 109 + 23771897 AAAGACCUUGUGCUGGUCAGUAAAGAAUGUAGAACCGGCCAAUGGCCAAUGGCCACUGGCCAAUGGGUAUGGCAACAAAAUGGCAUAGUUGAAGGUAAA----ACGCGAG-------CUC ....(((((..((((((((..............((((((((.(((((...))))).)))))....))).((....))...)))).))))..)))))...----.......-------... ( -34.00) >DroSec_CAF1 27535 102 + 1 AAAGACCUUGUGCUGGUCAGUAAAGGCUGUAGAACCGGCCAA-------UGGCCACUGGCCAAGGGGUAUGGGAACAAAAUGUUAUAGUUGAAGGUAAA----ACGAGAG-------CUC ....(((((....((((((((...((........))((((..-------.)))))))))))).))))).((....))....(((...(((........)----))...))-------).. ( -27.50) >DroSim_CAF1 28165 102 + 1 AAAGACCUUGUGCUGGUCAGUAAAGGCUGUAGAACCGGCCAA-------UGGCCACUGGCCAACGGGUAUGGGAACAAAAUGGCAUAGUUGAAGGUAAA----ACGAGAG-------CUC ....(((((..((((((((((...(((((......)))))..-------(((((...))))))).....((....))...)))).))))..)))))...----.......-------... ( -28.50) >DroAna_CAF1 19838 108 + 1 AAAGACCUUGUGCUGGUCAGUAAAGGU----AAACUGGCCAA-------UGGCCAUCGGCCG-UGGGUAUGGGAACAAAAUGGCAAAGUUGACCGUACACUUUACGUAAACAUUUUCCAC ......((..(((((((((((......----..))))))).(-------(((((...)))))-).))))..)).......(((.((((((...((((.....))))..))).))).))). ( -31.90) >consensus AAAGACCUUGUGCUGGUCAGUAAAGGCUGUAGAACCGGCCAA_______UGGCCACUGGCCAAUGGGUAUGGGAACAAAAUGGCAUAGUUGAAGGUAAA____ACGAGAG_______CUC ....(((((..((((((((((...((........))(((((........))))))))))))........((....)).........)))..)))))........................ (-23.98 = -24.60 + 0.62)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:34:29 2006