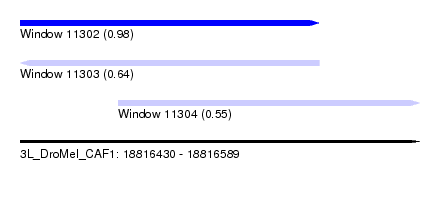
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 18,816,430 – 18,816,589 |
| Length | 159 |
| Max. P | 0.984144 |

| Location | 18,816,430 – 18,816,549 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.44 |
| Mean single sequence MFE | -31.93 |
| Consensus MFE | -30.43 |
| Energy contribution | -31.10 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.85 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.97 |
| SVM RNA-class probability | 0.984144 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
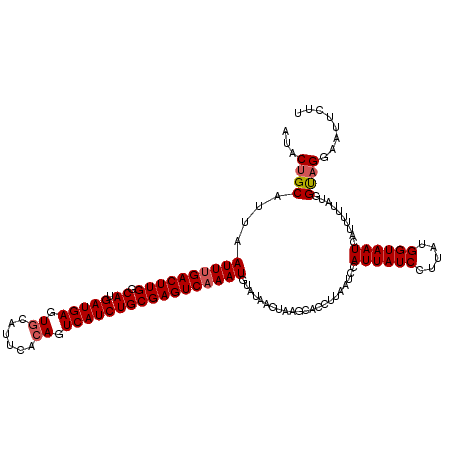
>3L_DroMel_CAF1 18816430 119 + 23771897 UACAA-AAGGGUUAGUUCCUAAGCUGUUCGAAUGCAACACAUACUGCAUUAAUUCGACUUGGCAUGAUGAGUGUAUACGAAGUCAUCUGCGAGUCAAAUGUAUAACUAAGCACCUUAAUC .....-(((((((((((.............((((((........)))))).....((((((.((.(((((.(........).)))))))))))))........)))).))).)))).... ( -28.00) >DroSec_CAF1 23904 120 + 1 AACAAAAAGGGUUAGUUCCAAAGCUGUUCGAAUGCAACACAUACUGCAUUUAUUUGACUUGGCAUGAUGAGUGCAUUCACAGUCAUCUGCGAGUCAAAUGUAUAACUAAGCACCUUAAUC ......(((((((((((............(((((((........)))))))((((((((((.((.(((((.((......)).)))))))))))))))))....)))).))).)))).... ( -34.60) >DroSim_CAF1 24183 119 + 1 AACAAAAAGGGUUAGUUCCUAAGCUGUUCGAAUGCAACACA-ACUGCAUUAAUUUGACUUGGCAUGAUGAGUGCAUUCACAGUCAUCUGCGAGUCAAAUGUAUAACUAAGCACCUUAAUC ......(((((((((((.............((((((.....-..)))))).((((((((((.((.(((((.((......)).)))))))))))))))))....)))).))).)))).... ( -33.20) >consensus AACAAAAAGGGUUAGUUCCUAAGCUGUUCGAAUGCAACACAUACUGCAUUAAUUUGACUUGGCAUGAUGAGUGCAUUCACAGUCAUCUGCGAGUCAAAUGUAUAACUAAGCACCUUAAUC ......(((((((((((.............((((((........)))))).((((((((((.((.(((((.((......)).)))))))))))))))))....)))).))).)))).... (-30.43 = -31.10 + 0.67)


| Location | 18,816,430 – 18,816,549 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.44 |
| Mean single sequence MFE | -29.43 |
| Consensus MFE | -28.72 |
| Energy contribution | -29.17 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.635794 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18816430 119 - 23771897 GAUUAAGGUGCUUAGUUAUACAUUUGACUCGCAGAUGACUUCGUAUACACUCAUCAUGCCAAGUCGAAUUAAUGCAGUAUGUGUUGCAUUCGAACAGCUUAGGAACUAACCCUU-UUGUA ....((((.(.((((((...(((((((((.((((((((............))))).)))..)))))))).(((((((......)))))))...........).)))))))))))-..... ( -26.10) >DroSec_CAF1 23904 120 - 1 GAUUAAGGUGCUUAGUUAUACAUUUGACUCGCAGAUGACUGUGAAUGCACUCAUCAUGCCAAGUCAAAUAAAUGCAGUAUGUGUUGCAUUCGAACAGCUUUGGAACUAACCCUUUUUGUU ....((((.(.((((((....((((((((.((((((((.(((....))).))))).)))..)))))))).(((((((......))))))).............)))))))))))...... ( -31.50) >DroSim_CAF1 24183 119 - 1 GAUUAAGGUGCUUAGUUAUACAUUUGACUCGCAGAUGACUGUGAAUGCACUCAUCAUGCCAAGUCAAAUUAAUGCAGU-UGUGUUGCAUUCGAACAGCUUAGGAACUAACCCUUUUUGUU ....((((.(.((((((...(((((((((.((((((((.(((....))).))))).)))..)))))))).(((((((.-....)))))))...........).)))))))))))...... ( -30.70) >consensus GAUUAAGGUGCUUAGUUAUACAUUUGACUCGCAGAUGACUGUGAAUGCACUCAUCAUGCCAAGUCAAAUUAAUGCAGUAUGUGUUGCAUUCGAACAGCUUAGGAACUAACCCUUUUUGUU ....((((.(.((((((...(((((((((.((((((((.(((....))).))))).)))..)))))))).(((((((......)))))))...........).)))))))))))...... (-28.72 = -29.17 + 0.45)

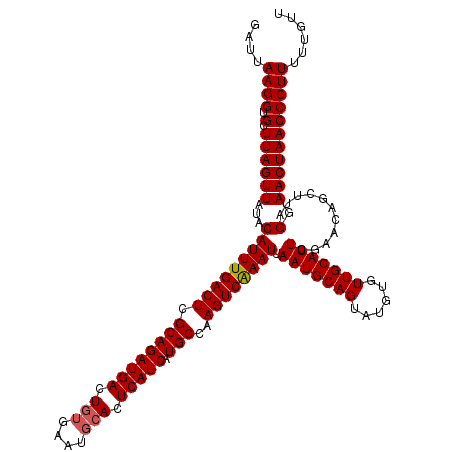
| Location | 18,816,469 – 18,816,589 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.67 |
| Mean single sequence MFE | -27.27 |
| Consensus MFE | -23.92 |
| Energy contribution | -24.70 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.549841 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18816469 120 + 23771897 AUACUGCAUUAAUUCGACUUGGCAUGAUGAGUGUAUACGAAGUCAUCUGCGAGUCAAAUGUAUAACUAAGCACCUUAAUCCAUUAUCCUUAUGGUAAUCAUUUUCAUGGCAGGACGUCUA ...((((........((((((.((.(((((.(........).)))))))))))))........................((((.......))))..............))))........ ( -24.20) >DroSec_CAF1 23944 120 + 1 AUACUGCAUUUAUUUGACUUGGCAUGAUGAGUGCAUUCACAGUCAUCUGCGAGUCAAAUGUAUAACUAAGCACCUUAAUCCAUUAUCCUUAUGGUAAUAAUUUUUAUGGUAGGAAUUCUU ..........(((((((((((.((.(((((.((......)).)))))))))))))))))).........................((((.((.((((......)))).))))))...... ( -28.90) >DroSim_CAF1 24223 119 + 1 A-ACUGCAUUAAUUUGACUUGGCAUGAUGAGUGCAUUCACAGUCAUCUGCGAGUCAAAUGUAUAACUAAGCACCUUAAUCCAUUAUCCUUAAGGUAAUCAUUUUUAUGGUCGGAAUUCUU .-.(((.((((((((((((((.((.(((((.((......)).)))))))))))))))))............(((((((..........)))))))...........)))))))....... ( -28.70) >consensus AUACUGCAUUAAUUUGACUUGGCAUGAUGAGUGCAUUCACAGUCAUCUGCGAGUCAAAUGUAUAACUAAGCACCUUAAUCCAUUAUCCUUAUGGUAAUCAUUUUUAUGGUAGGAAUUCUU ...((((....((((((((((.((.(((((.((......)).)))))))))))))))))......................((((((.....))))))..........))))........ (-23.92 = -24.70 + 0.78)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:34:25 2006