
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 18,811,179 – 18,811,371 |
| Length | 192 |
| Max. P | 0.734721 |

| Location | 18,811,179 – 18,811,279 |
|---|---|
| Length | 100 |
| Sequences | 3 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 95.33 |
| Mean single sequence MFE | -30.30 |
| Consensus MFE | -26.46 |
| Energy contribution | -27.13 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.734721 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
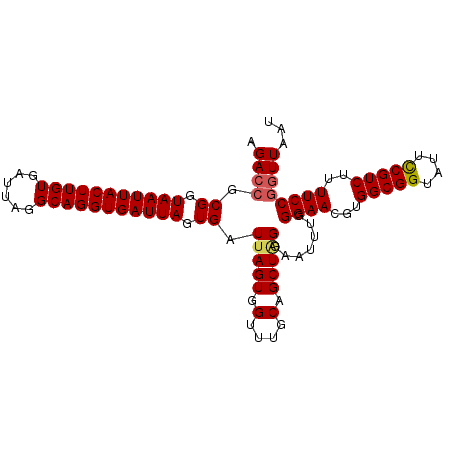
>3L_DroMel_CAF1 18811179 100 - 23771897 AGAAAGCGGUAAUUACUUGUGAUUAGGCAGGUGAUUAGUGAUAAGUGGUUUGCAGCUAGAAAUUUGGGAACGUGGCGGCAUUUCGUCUUUUCCGGUUAAU ....(((.((((((((((((.(((((........))))).)))))))).)))).)))......(((((((...(((((....))))).)))))))..... ( -25.30) >DroSec_CAF1 14966 100 - 1 AGACCGCGGUAAUUACUUGUGAUUAGGCAGGUGAUUAGUGAUUAGUGGUUUGCAGCUAGGAAUUUGGGAACGUGGCGGUAUUCCGUCUUUUCCGGUUAAU .((((.((.(((((((((((......))))))))))).)).(((((.(....).))))).......((((...(((((....)))))..))))))))... ( -31.60) >DroSim_CAF1 15237 100 - 1 AGACCGCGGUAAUUACUUGUGAUUAGGCAGGUGAUUAGUGAUUAGUGGUUUGCAGCUGGGAAUUUGGGAACGUGGCGGUAUUCCGUCUUUUCCGGUUAAU ((((((((.(((((((((((......))))))))))).).....)))))))..(((((((((...(....)..(((((....))))).)))))))))... ( -34.00) >consensus AGACCGCGGUAAUUACUUGUGAUUAGGCAGGUGAUUAGUGAUUAGUGGUUUGCAGCUAGGAAUUUGGGAACGUGGCGGUAUUCCGUCUUUUCCGGUUAAU .((((.((.(((((((((((......))))))))))).)).(((((.(....).))))).......((((...(((((....)))))..))))))))... (-26.46 = -27.13 + 0.67)

| Location | 18,811,279 – 18,811,371 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 81.91 |
| Mean single sequence MFE | -16.82 |
| Consensus MFE | -12.02 |
| Energy contribution | -13.65 |
| Covariance contribution | 1.63 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.71 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.528689 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
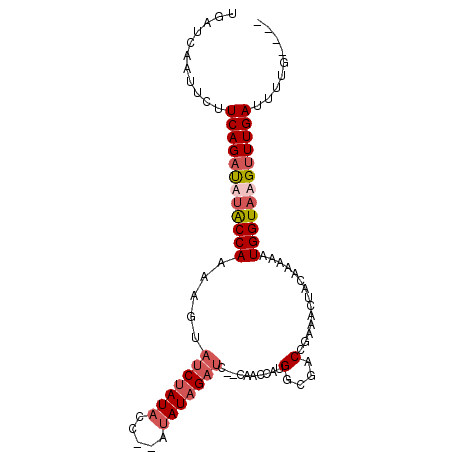
>3L_DroMel_CAF1 18811279 92 - 23771897 UGAUCAAUUCUUCAGAUAUACCAAUAGUAUCUAUACC--AUAUAGAUCCCCAACCAUGGCGACCGAAACUACAAAAAUGGUAAGUUUGAUUUUG---- ...........((((((.(((((......((((((..--.))))))((.(((....))).))...............))))).)))))).....---- ( -16.00) >DroSec_CAF1 15066 90 - 1 UGAUCAAUUCUUCAGAUUUACCAAAAGUAUCUAUACC--AUAUAGAUC--CAACCAUGGCGACCGAAACUACAAAAAUGGUAAGUUUGAUUUUG---- ...........((((((((((((.....(((((((..--.)))))))(--((....)))..................)))))))))))).....---- ( -18.00) >DroSim_CAF1 15337 90 - 1 UGAUCAAUUCUUCAGAUUUACCAAAAGUAUCUAUACC--AUAUAGAUC--CGACCAUGGCGACCGAAACUACAAAAAUGGUAAGUUUGAUUUUG---- ...........((((((((((((.....(((((((..--.))))))).--((.(....)))................)))))))))))).....---- ( -18.10) >DroAna_CAF1 12829 90 - 1 ACGCCAAUUCUUCAGACA-GCCACGA----CUAUUCCACAUAUUGAUC---AGCCAUGGCAACUGAAACAACGAAAAUGGUAAGAUUGAUUUUCAUCG ..........(((((...-((((.(.----((..((........))..---)).).))))..))))).....((((((.(......).)))))).... ( -15.20) >consensus UGAUCAAUUCUUCAGAUAUACCAAAAGUAUCUAUACC__AUAUAGAUC__CAACCAUGGCGACCGAAACUACAAAAAUGGUAAGUUUGAUUUUG____ ...........((((((((((((.....(((((((.....)))))))..........(....)..............))))))))))))......... (-12.02 = -13.65 + 1.63)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:34:19 2006