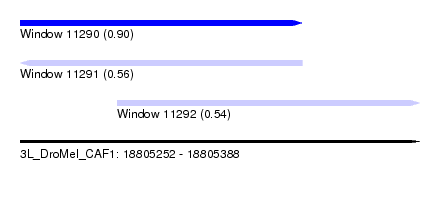
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 18,805,252 – 18,805,388 |
| Length | 136 |
| Max. P | 0.903420 |

| Location | 18,805,252 – 18,805,348 |
|---|---|
| Length | 96 |
| Sequences | 3 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 98.61 |
| Mean single sequence MFE | -29.61 |
| Consensus MFE | -26.64 |
| Energy contribution | -26.64 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.903420 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
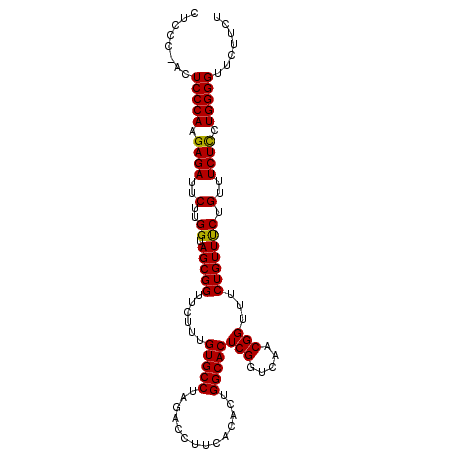
>3L_DroMel_CAF1 18805252 96 + 23771897 CAUCUUUGGUGCAAUGUUGUCAUUGUAAAGAGCCUCCCUACUCCCAAGAGAUUCUUGGUAGCGGUUCUUUGUGCCUAGACCUUCACACUGGCACUC ..........((((((....))))))((((((((...(((...(((((.....)))))))).))))))))(((((..............))))).. ( -27.34) >DroSec_CAF1 9550 95 + 1 CAUCUUUGGUGCAAUGUUGUCAUUGUAAAGAGCCUCCC-ACUCCCAAGAGAUUCUUGGGAGCGGUUCUUUGUGCCUAGACCUUCACACUGGCACUC ..........((((((....))))))((((((((....-.((((((((.....)))))))).))))))))(((((..............))))).. ( -34.14) >DroSim_CAF1 9492 95 + 1 CAUCUUUGGUGCAAUGUUGUCAUUGUAAAGAGCCUCCC-ACUCCCAAGAGAUUCUUGGUAGCGGUUCUUUGUGCCUAGACCUUCACACUGGCACUC ..........((((((....))))))((((((((....-.((.(((((.....))))).)).))))))))(((((..............))))).. ( -27.34) >consensus CAUCUUUGGUGCAAUGUUGUCAUUGUAAAGAGCCUCCC_ACUCCCAAGAGAUUCUUGGUAGCGGUUCUUUGUGCCUAGACCUUCACACUGGCACUC ..........((((((....))))))((((((((......((.(((((.....))))).)).))))))))(((((..............))))).. (-26.64 = -26.64 + 0.00)

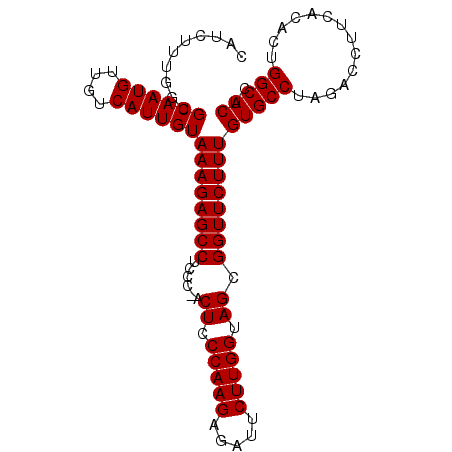
| Location | 18,805,252 – 18,805,348 |
|---|---|
| Length | 96 |
| Sequences | 3 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 98.61 |
| Mean single sequence MFE | -29.91 |
| Consensus MFE | -25.10 |
| Energy contribution | -25.10 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.555512 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18805252 96 - 23771897 GAGUGCCAGUGUGAAGGUCUAGGCACAAAGAACCGCUACCAAGAAUCUCUUGGGAGUAGGGAGGCUCUUUACAAUGACAACAUUGCACCAAAGAUG (.((((.(((((....(((..(((..........))).(((((.....)))))..((((((.....))))))...))).))))))))))....... ( -25.30) >DroSec_CAF1 9550 95 - 1 GAGUGCCAGUGUGAAGGUCUAGGCACAAAGAACCGCUCCCAAGAAUCUCUUGGGAGU-GGGAGGCUCUUUACAAUGACAACAUUGCACCAAAGAUG (.((((....((((((((((............(((((((((((.....)))))))))-)).)))).))))))((((....)))))))))....... ( -35.62) >DroSim_CAF1 9492 95 - 1 GAGUGCCAGUGUGAAGGUCUAGGCACAAAGAACCGCUACCAAGAAUCUCUUGGGAGU-GGGAGGCUCUUUACAAUGACAACAUUGCACCAAAGAUG (.((((....((((((((((............(((((.(((((.....))))).)))-)).)))).))))))((((....)))))))))....... ( -28.82) >consensus GAGUGCCAGUGUGAAGGUCUAGGCACAAAGAACCGCUACCAAGAAUCUCUUGGGAGU_GGGAGGCUCUUUACAAUGACAACAUUGCACCAAAGAUG (.((((.(((((....(((...(...(((((.(((((.(((((.....))))).))).....)).))))).)...))).))))))))))....... (-25.10 = -25.10 + -0.00)


| Location | 18,805,285 – 18,805,388 |
|---|---|
| Length | 103 |
| Sequences | 3 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 97.40 |
| Mean single sequence MFE | -27.44 |
| Consensus MFE | -25.78 |
| Energy contribution | -25.34 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.537715 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18805285 103 + 23771897 CUCCCUACUCCCAAGAGAUUCUUGGUAGCGGUUCUUUGUGCCUAGACCUUCACACUGGCACUCGGUCAACGGUUUCUGUUCCUGUUUCUUCUGGGGUUCUUCU ........(((((..(((..(..((.(((((......(((((..............)))))(((.....)))...))))))).)..)))..)))))....... ( -26.74) >DroSec_CAF1 9583 102 + 1 CUCCC-ACUCCCAAGAGAUUCUUGGGAGCGGUUCUUUGUGCCUAGACCUUCACACUGGCACUCGGUCAACGGUUUCUGUUUCUGUUUCUCCUGGGGUUCUUCU .((((-(((((((((.....)))))))).........(((((..............)))))..((..(((((.........)))))...)))))))....... ( -30.64) >DroSim_CAF1 9525 102 + 1 CUCCC-ACUCCCAAGAGAUUCUUGGUAGCGGUUCUUUGUGCCUAGACCUUCACACUGGCACUCGGUCAACGGUUUCUGUUUCUGUUUCUCCUGGGGUUCUUCU .....-..(((((.((((..(.(((.(((.(((((..(((((..............)))))..))..))).))).))).....)..)))).)))))....... ( -24.94) >consensus CUCCC_ACUCCCAAGAGAUUCUUGGUAGCGGUUCUUUGUGCCUAGACCUUCACACUGGCACUCGGUCAACGGUUUCUGUUUCUGUUUCUCCUGGGGUUCUUCU ........(((((.((((..(..((.(((((......(((((..............)))))(((.....)))...))))))).)..)))).)))))....... (-25.78 = -25.34 + -0.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:34:14 2006