| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 18,795,895 – 18,796,011 |
| Length | 116 |
| Max. P | 0.608298 |

| Location | 18,795,895 – 18,796,011 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 77.24 |
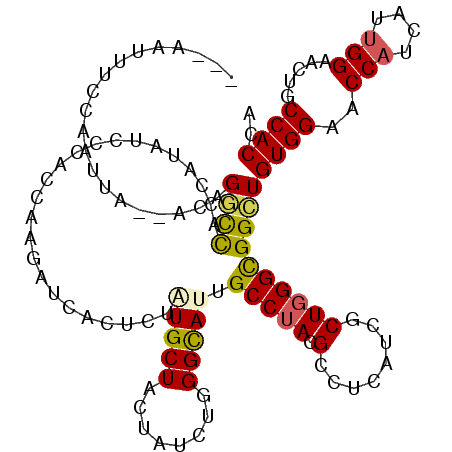
| Mean single sequence MFE | -31.27 |
| Consensus MFE | -17.80 |
| Energy contribution | -17.58 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.608298 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
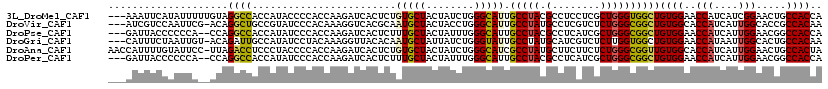
>3L_DroMel_CAF1 18795895 116 - 23771897 ---AAAUUCAUAUUUUUGUAGGCCACCAUACCCCACCAAGAUCACUCUGUGCUACUAUCUGGGCAUUGCCUACGCCUCCUCGCUGGGUGGCUGUGGAACCAUCAUCGGAACUGCCACCA ---..............((((((...............(((....)))(((((........))))).))))))((......))..((((((.((....((......)).)).)))))). ( -28.40) >DroVir_CAF1 1431 115 - 1 ---AUCGUCCAAUUCG-ACAGGCUGCCGUAUCCCACAAAGGUCACGCAAUGCUACUACCUGGGCAUUGCCUAUGCCUCGUCUCUGGGCGGCUGUGGCACCAUCAUUGGCACCGCCACAA ---.(((((((....(-((((((....((..((......))..))((((((((........))))))))....)))).)))..))))))).((((((.(((....)))....)))))). ( -42.50) >DroPse_CAF1 4683 114 - 1 ---GAUUACCCCCCA--CCAGGCCACCAUAUCCCACCAAGAUCACUCUUUGCUACUAUUUGGGCAUUGCCUACGCCUCAUCGCUGGGCGGCUGUGGAACCAUCAUUGGAACGGCCACCA ---............--...((((.(((((.((................((((........))))..(((((.((......))))))))).)))))..(((....)))...)))).... ( -31.50) >DroGri_CAF1 1938 115 - 1 ---CAUUUCUAAUUGU-ACAGAUUGCCAUAUCCUACAAAGGUUACACAAUGCUAUUAUCUGGGUAUUGCCUAUGCAUCGUCUCUUGGUGGCUGUGGAACCAUAAUUGGCACUGCCACAA ---.........((((-.(((..(((((...........((((.((((..(((((((...(((...(((....)))....))).))))))))))).)))).....))))))))..)))) ( -26.39) >DroAna_CAF1 1135 118 - 1 AACCAUUUUGUAUUCC-UUAGACCUCCCUACCCCACCAAGAUCACUCUGUGCUACUAUCUGGGCAUCGCCUAUGCUUCUUCUCUGGGCGGUUGUGGCACCAUCAUUGGAACUGCCACUA ................-...((((.(((.....(((..((....))..)))........(((((...)))))............))).))))(((((((((....)))...)))))).. ( -27.30) >DroPer_CAF1 4747 114 - 1 ---GAUUACCCCCCA--CCAGGCCACCAUAUCCCACCAAGAUCACUCUUUGCUACUAUUUGGGCAUUGCCUACGCCUCAUCGCUGGGCGGCUGUGGAACCAUCAUUGGAACGGCCACCA ---............--...((((.(((((.((................((((........))))..(((((.((......))))))))).)))))..(((....)))...)))).... ( -31.50) >consensus ___AAUUUCCAAUUA__ACAGGCCACCAUAUCCCACCAAGAUCACUCUAUGCUACUAUCUGGGCAUUGCCUACGCCUCAUCGCUGGGCGGCUGUGGAACCAUCAUUGGAACUGCCACCA ....................((((........................(((((........))))).(((((.(........))))))))))((((..(((....))).....)))).. (-17.80 = -17.58 + -0.22)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:34:06 2006