| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 18,707,934 – 18,708,036 |
| Length | 102 |
| Max. P | 0.885889 |

| Location | 18,707,934 – 18,708,036 |
|---|---|
| Length | 102 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.10 |
| Mean single sequence MFE | -31.23 |
| Consensus MFE | -13.32 |
| Energy contribution | -17.43 |
| Covariance contribution | 4.11 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.05 |
| Structure conservation index | 0.43 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.735939 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18707934 102 + 23771897 UAGUAGUUCACCCACAUA------AGCUGCGAGGGACACGUGAGGUUUCCCAGCGACA--AAAGUACGAAAUAUGUGGGCAUAAUAUACAUAUUGUAGAA---AUAUGU-------AUAU ..........((((((((------.((((.(.(..((.(....)))..)))))).((.--...))......))))))))....(((((((((((.....)---))))))-------)))) ( -33.40) >DroSec_CAF1 37385 94 + 1 UUGUAGUUCACCCACAUA------AGCUGCGAGGGACACGUGAGGUUUCCCAGCGACA--AAAGUACGAAAUAUGUGGGCAUAAUAUACAUAUUGUAC------------------AUAU .((((.....((((((((------.((((.(.(..((.(....)))..)))))).((.--...))......)))))))).......))))........------------------.... ( -26.00) >DroAna_CAF1 36072 117 + 1 UUGUAGUUCACCCACAUACUCGUGAGCUGUGAGGGACACGUGAGGUUUCUCAGCGACAGAAAAAUACAAAAUUUCUGUGAAA---GAACAUAUAGUAAGGCCUGUAUGUAUGGUACGUAU ...((((((((..........))))))))((((..((.(....)))..))))((((((((((.........)))))))....---..((((((((......))))))))......))).. ( -34.30) >consensus UUGUAGUUCACCCACAUA______AGCUGCGAGGGACACGUGAGGUUUCCCAGCGACA__AAAGUACGAAAUAUGUGGGCAUAAUAUACAUAUUGUAA______UAUGU_______AUAU ..........((((((((.......((((.(.(..((.(....)))..)))))).((......))......))))))))......................................... (-13.32 = -17.43 + 4.11)

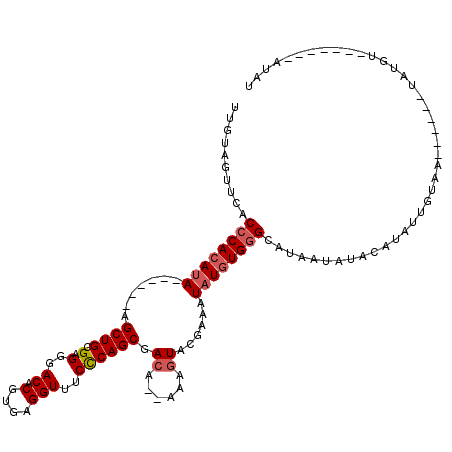
| Location | 18,707,934 – 18,708,036 |
|---|---|
| Length | 102 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.10 |
| Mean single sequence MFE | -26.77 |
| Consensus MFE | -12.86 |
| Energy contribution | -13.53 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.48 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.885889 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18707934 102 - 23771897 AUAU-------ACAUAU---UUCUACAAUAUGUAUAUUAUGCCCACAUAUUUCGUACUUU--UGUCGCUGGGAAACCUCACGUGUCCCUCGCAGCU------UAUGUGGGUGAACUACUA ((((-------((((((---(.....)))))))))))...(((((((((....(.((...--.)))(((((....))....(((.....)))))).------)))))))))......... ( -29.80) >DroSec_CAF1 37385 94 - 1 AUAU------------------GUACAAUAUGUAUAUUAUGCCCACAUAUUUCGUACUUU--UGUCGCUGGGAAACCUCACGUGUCCCUCGCAGCU------UAUGUGGGUGAACUACAA ..((------------------((((.....))))))...(((((((((....(.((...--.)))(((((....))....(((.....)))))).------)))))))))......... ( -25.20) >DroAna_CAF1 36072 117 - 1 AUACGUACCAUACAUACAGGCCUUACUAUAUGUUC---UUUCACAGAAAUUUUGUAUUUUUCUGUCGCUGAGAAACCUCACGUGUCCCUCACAGCUCACGAGUAUGUGGGUGAACUACAA .(((((......((((.((......)).))))(((---((..(((((((.........)))))))....))))).....)))))((.(((((((((....))).)))))).))....... ( -25.30) >consensus AUAU_______ACAUA______CUACAAUAUGUAUAUUAUGCCCACAUAUUUCGUACUUU__UGUCGCUGGGAAACCUCACGUGUCCCUCGCAGCU______UAUGUGGGUGAACUACAA .......................................((((((((((....(.((......)))((((((....)))..(((.....)))))).......))))))))))........ (-12.86 = -13.53 + 0.67)

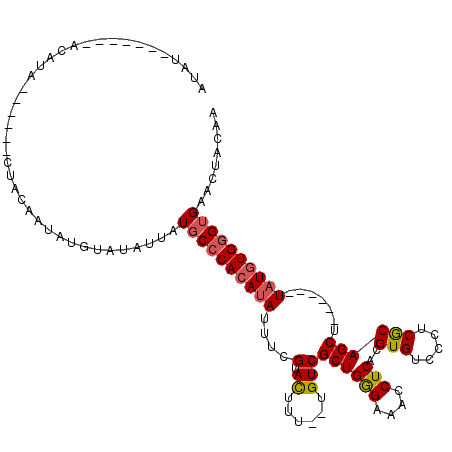
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:33:49 2006