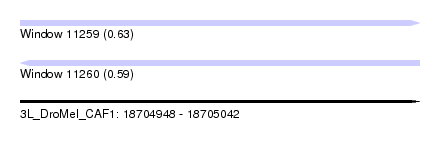
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 18,704,948 – 18,705,042 |
| Length | 94 |
| Max. P | 0.634495 |

| Location | 18,704,948 – 18,705,042 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 76.96 |
| Mean single sequence MFE | -29.35 |
| Consensus MFE | -14.91 |
| Energy contribution | -17.41 |
| Covariance contribution | 2.50 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.634495 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18704948 94 + 23771897 AAGCUUCGCUG-----------UGCCGCUUCAAUAAAGCGCUUGUAC-------GGAAAAAACAAAACUUUCCCACGAGCCAAGCAUUUUAGAGCAGGGAUUCCGUGCGAAA ..((((.((((-----------((((((((.....)))))...))))-------(((((.........)))))....))).))))........(((.((...)).))).... ( -25.00) >DroSec_CAF1 34370 105 + 1 AAGCUUCGCACAGCUCUUCGCCAGCCGCUUCAAUAAAGCGCUUGGAC-------GGAAAAAACAAAACUUUCCCACGAGCCAAGCAUUUUAGAGCGGGGAUUCCGUGCGAAA ....(((((((.(.((((((((....((((.....))))((((((..-------(((((.........)))))......))))))......).)))))))..).))))))). ( -34.70) >DroAna_CAF1 33336 111 + 1 AAGCCUUCUUGAGCUCUACGCCACCAAGUUCAAUAAUAUUGUUUAACAAGUCGGGCAAAAAACAAAACUUUCACACGAGCCAACAAUUUUUGAGCGGGUAAUCCC-CCGAAA ..((((..(((((((...........))))))).....(((.....)))...)))).............(((......(((((......))).))(((......)-))))). ( -18.30) >DroSim_CAF1 35630 105 + 1 AAGCUUCGCACAGCUCUUCGCCAGCCGCUUCAAUAAAGCGCUUGGAC-------GGGAAAAACAAAACUUUCCCACGAGCCAAGCAUUUUAGAGCGGGGAUUCCGUGCGAAA ....(((((((.(.((((((((....((((.....))))((((((..-------((((((........)))))).....))))))......).)))))))..).))))))). ( -39.40) >consensus AAGCUUCGCACAGCUCUUCGCCAGCCGCUUCAAUAAAGCGCUUGGAC_______GGAAAAAACAAAACUUUCCCACGAGCCAAGCAUUUUAGAGCGGGGAUUCCGUGCGAAA ....(((((((.((((..........((((.....))))((((((.........(((((.........)))))......))))))......)))).((....))))))))). (-14.91 = -17.41 + 2.50)

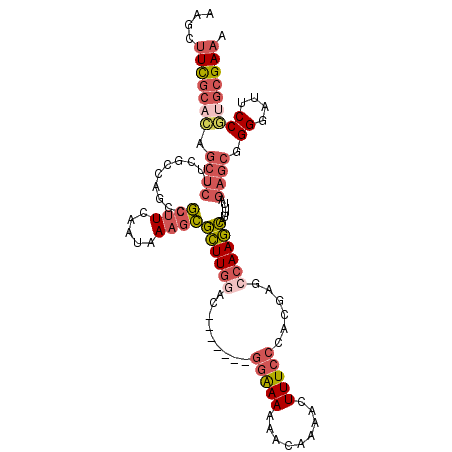
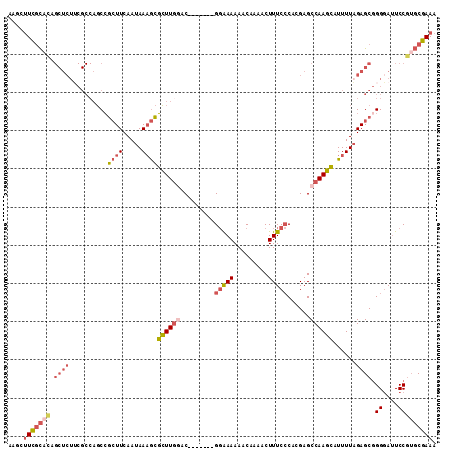
| Location | 18,704,948 – 18,705,042 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 76.96 |
| Mean single sequence MFE | -33.40 |
| Consensus MFE | -20.10 |
| Energy contribution | -20.91 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.590188 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18704948 94 - 23771897 UUUCGCACGGAAUCCCUGCUCUAAAAUGCUUGGCUCGUGGGAAAGUUUUGUUUUUUCC-------GUACAAGCGCUUUAUUGAAGCGGCA-----------CAGCGAAGCUU ....(((.((....)))))........((((.(((.(((((((((.......))))))-------.......((((((...)))))).))-----------)))).)))).. ( -28.50) >DroSec_CAF1 34370 105 - 1 UUUCGCACGGAAUCCCCGCUCUAAAAUGCUUGGCUCGUGGGAAAGUUUUGUUUUUUCC-------GUCCAAGCGCUUUAUUGAAGCGGCUGGCGAAGAGCUGUGCGAAGCUU ((((((((((..((((.(((.......((((((....((((((((......)))))))-------).))))))(((((...)))))))).)).))....))))))))))... ( -39.10) >DroAna_CAF1 33336 111 - 1 UUUCGG-GGGAUUACCCGCUCAAAAAUUGUUGGCUCGUGUGAAAGUUUUGUUUUUUGCCCGACUUGUUAAACAAUAUUAUUGAACUUGGUGGCGUAGAGCUCAAGAAGGCUU ....((-(......)))((((((..((((((((((((.(..((((......))))..).)))...))).))))))....)))).((((..(((.....)))))))...)).. ( -24.00) >DroSim_CAF1 35630 105 - 1 UUUCGCACGGAAUCCCCGCUCUAAAAUGCUUGGCUCGUGGGAAAGUUUUGUUUUUCCC-------GUCCAAGCGCUUUAUUGAAGCGGCUGGCGAAGAGCUGUGCGAAGCUU ((((((((((..((((.(((.......((((((....((((((((......)))))))-------).))))))(((((...)))))))).)).))....))))))))))... ( -42.00) >consensus UUUCGCACGGAAUCCCCGCUCUAAAAUGCUUGGCUCGUGGGAAAGUUUUGUUUUUUCC_______GUCCAAGCGCUUUAUUGAAGCGGCUGGCGAAGAGCUCAGCGAAGCUU ((((((((.......((((((((((.(((((((.....(((((((......))))))).........))))))).))))..).)))))..(((.....)))))))))))... (-20.10 = -20.91 + 0.81)

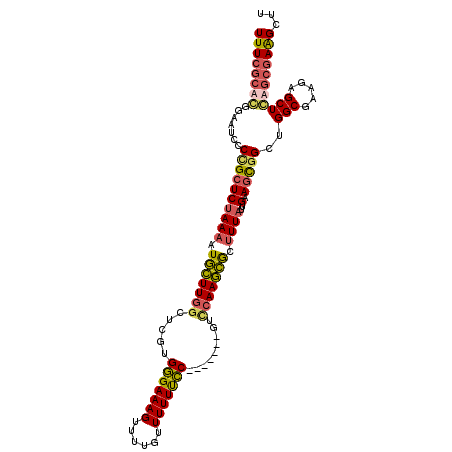
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:33:43 2006