
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 18,704,415 – 18,704,670 |
| Length | 255 |
| Max. P | 0.973488 |

| Location | 18,704,415 – 18,704,516 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.68 |
| Mean single sequence MFE | -12.35 |
| Consensus MFE | -10.84 |
| Energy contribution | -11.08 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.88 |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.532184 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18704415 101 - 23771897 AUUGCUUAUCUGUGCAUUAAGAUUUGCAAUUCCAUUACAUUAAUUACAUAAUUGAAAUGGCAACCAUAAUAUUUUCACCCCAACUACAUUCCAUUAUUUUG------------------- (((((..((((........))))..))))).(((((...((((((....))))))))))).........................................------------------- ( -12.00) >DroSec_CAF1 33826 101 - 1 AUUGCUUAUCUGUGCGUUAAGAUUUGCAAUUCCAUUACAUUAAUUACAUAAUUGAAAUGGCAACCAUAAUAUUUUCACCCCUACUACAUUCCAUUAUUUUC------------------- (((((..((((........))))..))))).(((((...((((((....))))))))))).........................................------------------- ( -12.00) >DroAna_CAF1 32921 114 - 1 AUUGCUUAUCUGUGCAUUAAGAUUUGCAUUUCCAUUACAUUAAUUAUAUAAUUGGACUGGUAAACAUUACUAUGCCACCU------CAUCCCUUUAUCCCCGUCACCCUCCGAAAUAGCC ...((..((((........))))..))(((((...............((((..(((..(((...(((....)))..))).------..)))..))))..............))))).... ( -13.75) >DroSim_CAF1 35069 101 - 1 AUUGCUUAUCUGUGCAUUAAGAUUUGCAAUUCCAUUACAUUAAUUACAUAAUUGAAAUGGCAACCAUAAUAUUUUCACCCCUACUACAUUCCAUUAUUUUC------------------- (((((..((((........))))..))))).(((((...((((((....))))))))))).........................................------------------- ( -12.00) >DroYak_CAF1 35361 101 - 1 AUUGCUUAUCUGUGCAUUAAGAUUUGCAAUUCCAUUACAUUAAUUACAUAAUUGAAAUGGCAAACAUCAUACUUUCACCCUUACUGCAUUCCAUUAUUUUG------------------- (((((..((((........))))..))))).(((((...((((((....))))))))))).........................................------------------- ( -12.00) >consensus AUUGCUUAUCUGUGCAUUAAGAUUUGCAAUUCCAUUACAUUAAUUACAUAAUUGAAAUGGCAACCAUAAUAUUUUCACCCCUACUACAUUCCAUUAUUUUC___________________ (((((..((((........))))..))))).(((((...((((((....)))))))))))............................................................ (-10.84 = -11.08 + 0.24)


| Location | 18,704,436 – 18,704,554 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.92 |
| Mean single sequence MFE | -21.77 |
| Consensus MFE | -18.20 |
| Energy contribution | -18.40 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.71 |
| SVM RNA-class probability | 0.973488 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18704436 118 - 23771897 UAUUCAC-UUAUUUGAAGAGGGA-AUAAUUUGCCAUUAUAAUUGCUUAUCUGUGCAUUAAGAUUUGCAAUUCCAUUACAUUAAUUACAUAAUUGAAAUGGCAACCAUAAUAUUUUCACCC (((((.(-((.......))).))-)))..((((((((..((((((..((((........))))..))))))........((((((....))))))))))))))................. ( -23.50) >DroSec_CAF1 33847 118 - 1 UAUUCAC-UUAUUUGAAGAGGGA-AUAAUUUGCCAUUAUAAUUGCUUAUCUGUGCGUUAAGAUUUGCAAUUCCAUUACAUUAAUUACAUAAUUGAAAUGGCAACCAUAAUAUUUUCACCC (((((.(-((.......))).))-)))..((((((((..((((((..((((........))))..))))))........((((((....))))))))))))))................. ( -23.50) >DroAna_CAF1 32955 118 - 1 --UUCAAUUGAAUUGAAGACCCAAAUAAUUUGCUUUUAUAAUUGCUUAUCUGUGCAUUAAGAUUUGCAUUUCCAUUACAUUAAUUAUAUAAUUGGACUGGUAAACAUUACUAUGCCACCU --((((((((..(((......)))((((((............(((..((((........))))..))).............)))))).)))))))).(((((..........)))))... ( -15.71) >DroSim_CAF1 35090 118 - 1 UAUUCAC-UUAUUUGAAGAGGGA-AUAAUUUGCCAUUAUAAUUGCUUAUCUGUGCAUUAAGAUUUGCAAUUCCAUUACAUUAAUUACAUAAUUGAAAUGGCAACCAUAAUAUUUUCACCC (((((.(-((.......))).))-)))..((((((((..((((((..((((........))))..))))))........((((((....))))))))))))))................. ( -23.50) >DroYak_CAF1 35382 119 - 1 UAUUCAC-UUAUUUGAAGAGGGAAAUUAUUUGCUAUUAUAAUUGCUUAUCUGUGCAUUAAGAUUUGCAAUUCCAUUACAUUAAUUACAUAAUUGAAAUGGCAAACAUCAUACUUUCACCC ..((((.-.....))))(((((......(((((((((..((((((..((((........))))..))))))........((((((....))))))))))))))).......))))).... ( -22.62) >consensus UAUUCAC_UUAUUUGAAGAGGGA_AUAAUUUGCCAUUAUAAUUGCUUAUCUGUGCAUUAAGAUUUGCAAUUCCAUUACAUUAAUUACAUAAUUGAAAUGGCAACCAUAAUAUUUUCACCC ..((((.......))))(((((.......((((((((..((((((..((((........))))..))))))........((((((....))))))))))))))........))))).... (-18.20 = -18.40 + 0.20)



| Location | 18,704,476 – 18,704,594 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 93.52 |
| Mean single sequence MFE | -21.43 |
| Consensus MFE | -18.85 |
| Energy contribution | -19.35 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.787896 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18704476 118 - 23771897 AUAGAAUGCACUUUCAAUGCAUUUAAACUUCCAUUUAUCCUAUUCACUUAUUUGAAGAGGGA-AUAAUUUGCCAUUAUAAUUGCUUAUCUGUGCAUUAAGAUUUGCAAUUCCAUUACAU ...(((((((.......))))))).............((((.((((......))))..))))-...............((((((..((((........))))..))))))......... ( -21.00) >DroSec_CAF1 33887 117 - 1 AUAGAAUGCAAU-UCAAUGCAUUUAAACUACCAUUUAUCCUAUUCACUUAUUUGAAGAGGGA-AUAAUUUGCCAUUAUAAUUGCUUAUCUGUGCGUUAAGAUUUGCAAUUCCAUUACAU ...(((((((..-....))))))).............((((.((((......))))..))))-...............((((((..((((........))))..))))))......... ( -21.70) >DroSim_CAF1 35130 117 - 1 AUAGAAUGCAGU-UGAAUGCAUUUAAACUACCAUUUAUCCUAUUCACUUAUUUGAAGAGGGA-AUAAUUUGCCAUUAUAAUUGCUUAUCUGUGCAUUAAGAUUUGCAAUUCCAUUACAU ...(((((((..-....))))))).............((((.((((......))))..))))-...............((((((..((((........))))..))))))......... ( -22.50) >DroYak_CAF1 35422 119 - 1 AUAGAAUUAAAAUGCAAUGCAUUUAAACAUCAAUUUAUCCUAUUCACUUAUUUGAAGAGGGAAAUUAUUUGCUAUUAUAAUUGCUUAUCUGUGCAUUAAGAUUUGCAAUUCCAUUACAU ((((.....((((((...)))))).......(((((.((((.((((......)))).))))))))).....))))...((((((..((((........))))..))))))......... ( -20.50) >consensus AUAGAAUGCAAU_UCAAUGCAUUUAAACUACCAUUUAUCCUAUUCACUUAUUUGAAGAGGGA_AUAAUUUGCCAUUAUAAUUGCUUAUCUGUGCAUUAAGAUUUGCAAUUCCAUUACAU ...(((((((.......))))))).............((((.((((......)))).)))).................((((((..((((........))))..))))))......... (-18.85 = -19.35 + 0.50)


| Location | 18,704,516 – 18,704,634 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 93.10 |
| Mean single sequence MFE | -23.77 |
| Consensus MFE | -19.08 |
| Energy contribution | -19.82 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.563717 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18704516 118 + 23771897 UAUAAUGGCAAAUUAU-UCCCUCUUCAAAUAAGUGAAUAGGAUAAAUGGAAGUUUAAAUGCAUUGAAAGUGCAUUCUAUAAUAAGAAUGCUGAGCAGGUCAUAUUUAAUUGAAUGCAUU ..(((((.((......-..(((.((((......)))).))).(((((....)))))..))))))).....(((((((......)))))))...(((..(((........))).)))... ( -22.90) >DroSec_CAF1 33927 117 + 1 UAUAAUGGCAAAUUAU-UCCCUCUUCAAAUAAGUGAAUAGGAUAAAUGGUAGUUUAAAUGCAUUGA-AUUGCAUUCUAUAAUAAGAAUGCUGAGCAGGUCAUAUUUAAUUGAAUGCAUU .........(((((((-(.(((.((((......)))).)))......)))))))).((((((((.(-((((((((((......)))))))(((.....))).....)))).)))))))) ( -24.20) >DroSim_CAF1 35170 117 + 1 UAUAAUGGCAAAUUAU-UCCCUCUUCAAAUAAGUGAAUAGGAUAAAUGGUAGUUUAAAUGCAUUCA-ACUGCAUUCUAUAAUAAGAAUGCUGAGCAGGUCAUAUUUAAUUGAAUGCAUU .........(((((((-(.(((.((((......)))).)))......)))))))).((((((((((-(..(((((((......)))))))(((.....))).......))))))))))) ( -26.20) >DroYak_CAF1 35462 119 + 1 UAUAAUAGCAAAUAAUUUCCCUCUUCAAAUAAGUGAAUAGGAUAAAUUGAUGUUUAAAUGCAUUGCAUUUUAAUUCUAUACUAAGAAUGCUGAGCAGGUAAUAUUUAAUUGAAUGCAUU .......(((.........(((.((((......)))).)))...((((((...(((..(((...(((((((............)))))))...)))..)))...))))))...)))... ( -21.80) >consensus UAUAAUGGCAAAUUAU_UCCCUCUUCAAAUAAGUGAAUAGGAUAAAUGGAAGUUUAAAUGCAUUGA_AUUGCAUUCUAUAAUAAGAAUGCUGAGCAGGUCAUAUUUAAUUGAAUGCAUU .......(((.........(((.((((......)))).))).(((((....)))))..))).........(((((((......)))))))...(((..(((........))).)))... (-19.08 = -19.82 + 0.75)


| Location | 18,704,516 – 18,704,634 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 93.10 |
| Mean single sequence MFE | -23.17 |
| Consensus MFE | -17.67 |
| Energy contribution | -18.42 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.80 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.805119 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18704516 118 - 23771897 AAUGCAUUCAAUUAAAUAUGACCUGCUCAGCAUUCUUAUUAUAGAAUGCACUUUCAAUGCAUUUAAACUUCCAUUUAUCCUAUUCACUUAUUUGAAGAGGGA-AUAAUUUGCCAUUAUA ((((((((..........(((.....)))(((((((......)))))))......))))))))..............((((.((((......))))..))))-................ ( -21.60) >DroSec_CAF1 33927 117 - 1 AAUGCAUUCAAUUAAAUAUGACCUGCUCAGCAUUCUUAUUAUAGAAUGCAAU-UCAAUGCAUUUAAACUACCAUUUAUCCUAUUCACUUAUUUGAAGAGGGA-AUAAUUUGCCAUUAUA ((((((((.((((.....(((.....)))(((((((......))))))))))-).))))))))..............((((.((((......))))..))))-................ ( -23.40) >DroSim_CAF1 35170 117 - 1 AAUGCAUUCAAUUAAAUAUGACCUGCUCAGCAUUCUUAUUAUAGAAUGCAGU-UGAAUGCAUUUAAACUACCAUUUAUCCUAUUCACUUAUUUGAAGAGGGA-AUAAUUUGCCAUUAUA (((((((((((((.....(((.....)))(((((((......))))))))))-))))))))))..............((((.((((......))))..))))-................ ( -29.60) >DroYak_CAF1 35462 119 - 1 AAUGCAUUCAAUUAAAUAUUACCUGCUCAGCAUUCUUAGUAUAGAAUUAAAAUGCAAUGCAUUUAAACAUCAAUUUAUCCUAUUCACUUAUUUGAAGAGGGAAAUUAUUUGCUAUUAUA ...(((............(((..(((...(((((.(((((.....))))))))))...)))..))).....(((((.((((.((((......)))).)))))))))...)))....... ( -18.10) >consensus AAUGCAUUCAAUUAAAUAUGACCUGCUCAGCAUUCUUAUUAUAGAAUGCAAU_UCAAUGCAUUUAAACUACCAUUUAUCCUAUUCACUUAUUUGAAGAGGGA_AUAAUUUGCCAUUAUA ((((((((..........(((.....)))(((((((......)))))))......))))))))..............((((.((((......)))).)))).................. (-17.67 = -18.42 + 0.75)


| Location | 18,704,554 – 18,704,670 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.91 |
| Mean single sequence MFE | -24.38 |
| Consensus MFE | -16.27 |
| Energy contribution | -18.03 |
| Covariance contribution | 1.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.22 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.575053 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18704554 116 - 23771897 CUUUCAAAACAAAUA----CAUGUGACAAUGGUGCAAGGAAAUGCAUUCAAUUAAAUAUGACCUGCUCAGCAUUCUUAUUAUAGAAUGCACUUUCAAUGCAUUUAAACUUCCAUUUAUCC ...............----........(((((...(((.(((((((((..........(((.....)))(((((((......)))))))......)))))))))...))))))))..... ( -22.20) >DroSec_CAF1 33965 119 - 1 CUUUCAAAACAAAUACACACAUGUGACAAUGGUGCAAGGAAAUGCAUUCAAUUAAAUAUGACCUGCUCAGCAUUCUUAUUAUAGAAUGCAAU-UCAAUGCAUUUAAACUACCAUUUAUCC ...............(((....)))..(((((((.....(((((((((.((((.....(((.....)))(((((((......))))))))))-).)))))))))....)))))))..... ( -28.00) >DroSim_CAF1 35208 119 - 1 CUUUCAAAACAAAUACACACAUGUGACAAUGGUGCAAGGAAAUGCAUUCAAUUAAAUAUGACCUGCUCAGCAUUCUUAUUAUAGAAUGCAGU-UGAAUGCAUUUAAACUACCAUUUAUCC ...............(((....)))..(((((((.....((((((((((((((.....(((.....)))(((((((......))))))))))-)))))))))))....)))))))..... ( -34.20) >DroYak_CAF1 35501 116 - 1 UUUUCAAAGCAAAUA----CAUGUGACAAUGGUACAAGGAAAUGCAUUCAAUUAAAUAUUACCUGCUCAGCAUUCUUAGUAUAGAAUUAAAAUGCAAUGCAUUUAAACAUCAAUUUAUCC ........(((....----(((......)))...........((((((.((((..((((((..(((...)))....))))))..))))..)))))).))).................... ( -13.10) >consensus CUUUCAAAACAAAUA____CAUGUGACAAUGGUGCAAGGAAAUGCAUUCAAUUAAAUAUGACCUGCUCAGCAUUCUUAUUAUAGAAUGCAAU_UCAAUGCAUUUAAACUACCAUUUAUCC ...........................(((((((.....(((((((((..........(((.....)))(((((((......)))))))......)))))))))....)))))))..... (-16.27 = -18.03 + 1.75)

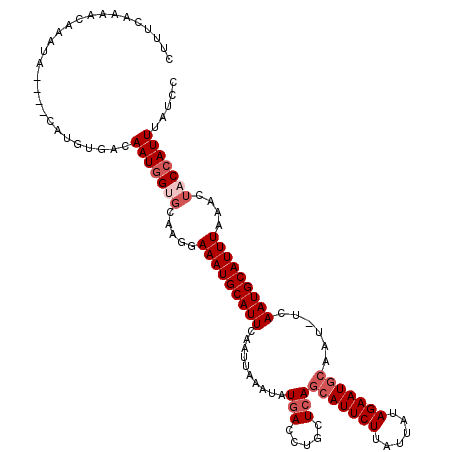
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:33:41 2006