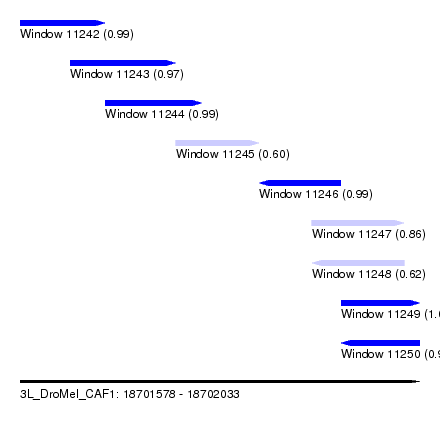
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 18,701,578 – 18,702,033 |
| Length | 455 |
| Max. P | 0.999433 |

| Location | 18,701,578 – 18,701,675 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.16 |
| Mean single sequence MFE | -26.28 |
| Consensus MFE | -15.86 |
| Energy contribution | -19.42 |
| Covariance contribution | 3.56 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.36 |
| Structure conservation index | 0.60 |
| SVM decision value | 2.51 |
| SVM RNA-class probability | 0.994787 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18701578 97 + 23771897 ------------AC-G----UAUAUAUUGUAUCUGCGUUUUCAUUUUCAUUUCCAUGGGAAUGGGAAU------GGCAGCAAACUGCUGACCAAAUCAAUUUCGCAAUGCAAAAUGAAAA ------------..-.----..(((.((((((.((((((..((((..(((....)))..))))..))(------((((((.....)))).))).........)))))))))).))).... ( -29.10) >DroSec_CAF1 31001 98 + 1 ------------AAUU----UAUAUAUUGUAUAUGCGUUUUCAUUUUCAUUUCCAUGGGAAUGGGAAU------GGCAGCAAACUGCUGACCAAAUCAAUUUCGCAAUGCAAAAUGAAAA ------------....----..(((.((((((.((((((..((((..(((....)))..))))..))(------((((((.....)))).))).........)))))))))).))).... ( -29.10) >DroAna_CAF1 30103 93 + 1 GGCAAGAA-------U----UAUAUGUAGUAC---------CAAUA-UAUAUAAAUGUACAUUGAAAU------UGCAACAAACUGCUGACCAAAUCAAUUUCGAAAUGUAAAAUGAAAA ........-------(----((((((((....---------...))-)))))))...(((((((((((------((((......))).((.....))))))))))..))))......... ( -12.70) >DroSim_CAF1 32175 98 + 1 ------------ACUU----UAUAUAUUGUAUAUGCGUUUUCAUUUUCAUUUCCAUGGGAAUGGGAAU------GGCAGCAAACUGCUGACCAAAUCAAUUUCGCAAUGCAAAAUGAAAA ------------....----..(((.((((((.((((((..((((..(((....)))..))))..))(------((((((.....)))).))).........)))))))))).))).... ( -29.10) >DroEre_CAF1 31562 103 + 1 GGCAUGAAAUACAC-G----UAUAUAUUGUAUAUGCAUUUCCAUUUUCAUUUCCAUGGGCAU------------GGCAGCAAACUGCUGACCAAAUCAAUUUCGCAAUGCAAAAUGAAAA .(((((((((....-(----((((((...)))))))...(((((..........)))))..(------------((((((.....)))).))).....)))))...)))).......... ( -20.70) >DroYak_CAF1 32419 119 + 1 GGCAUAAAAUACAC-GUUCAUAUAUAUUGUAUAUGCAUUUCCAUUUUCAUUUCCAUGGGAAUGGGAAUGGGAACGGCAGCAAACUGCUGACCAAAUCAAUUUCGCAAUGCAAAAUGAAAA ..............-.(((((.....((((((.(((.((((((((..((((((....))))))..)))))))).((((((.....)))).))...........))))))))).))))).. ( -37.00) >consensus ____________AC_G____UAUAUAUUGUAUAUGCGUUUUCAUUUUCAUUUCCAUGGGAAUGGGAAU______GGCAGCAAACUGCUGACCAAAUCAAUUUCGCAAUGCAAAAUGAAAA ......................(((.((((((.(((((((((((((((((....))))))))))))))......((((((.....)))).))...........))))))))).))).... (-15.86 = -19.42 + 3.56)

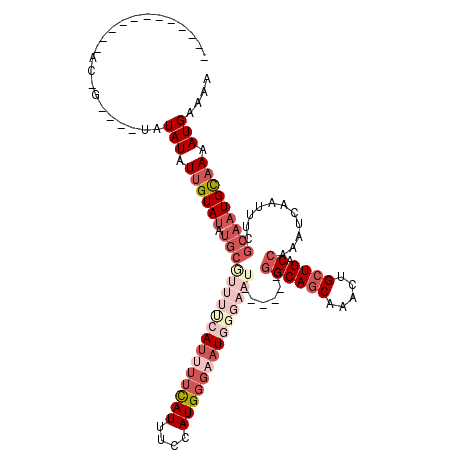

| Location | 18,701,635 – 18,701,755 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.17 |
| Mean single sequence MFE | -33.68 |
| Consensus MFE | -31.43 |
| Energy contribution | -31.43 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.966389 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18701635 120 + 23771897 AAACUGCUGACCAAAUCAAUUUCGCAAUGCAAAAUGAAAAUGUUACAAGGGAUUUUCGGGGAUCGAUGCGGACGGACUGCGGAAAUUUGUUGCCGCUGCUUAGAUGCCCGUAAAAUUGCA ....((.((((....(((.(((.((...))))).)))....)))))).(.((((((((((.(((...((((.(((...((((....))))..)))))))...))).)))).)))))).). ( -33.00) >DroSec_CAF1 31059 120 + 1 AAACUGCUGACCAAAUCAAUUUCGCAAUGCAAAAUGAAAAUGUUACAAGGGAUUUUCGGGGAUCGAUGCGGACGGACUGCGGAAAUUUGUUGCCGCUGCUUAGAUGCCCGUAAAAUUGCA ....((.((((....(((.(((.((...))))).)))....)))))).(.((((((((((.(((...((((.(((...((((....))))..)))))))...))).)))).)))))).). ( -33.00) >DroAna_CAF1 30156 120 + 1 AAACUGCUGACCAAAUCAAUUUCGAAAUGUAAAAUGAAAAUGUUAGAGGGGAUUUUCGGGGAUCGAUGCGGACGGACUGCGGAAAUUUGUUACCGCUGCUUAGAUGCCCGUAAAAUUGCA ...((.(((((....(((.(((........))).)))....)))))))(.((((((((((.(((...((((.(((...((((....))))..)))))))...))).)))).)))))).). ( -33.80) >DroSim_CAF1 32233 120 + 1 AAACUGCUGACCAAAUCAAUUUCGCAAUGCAAAAUGAAAAUGUUACAAGGGAUUUUCGGGGAUCGAUGCGGACGGACUGCGGAAAUUUGUUGCCGCUGCUUAGAUGCCCGUAAAAUUGCA ....((.((((....(((.(((.((...))))).)))....)))))).(.((((((((((.(((...((((.(((...((((....))))..)))))))...))).)))).)))))).). ( -33.00) >DroEre_CAF1 31625 120 + 1 AAACUGCUGACCAAAUCAAUUUCGCAAUGCAAAAUGAAAAUGUUACAGGGGAUUUUCGGGGAUCGAUGCGGACGGAAUGCGGAAAUUUGUUGCCGCUGCUUAGAUGCCCGUAAAAUUGCA ...(((.((((....(((.(((.((...))))).)))....)))))))(.((((((((((.(((...((((.(((...((((....))))..)))))))...))).)))).)))))).). ( -36.30) >DroYak_CAF1 32498 120 + 1 AAACUGCUGACCAAAUCAAUUUCGCAAUGCAAAAUGAAAAUGUUACAAGGGAUUUUCGGGGAUCGAUGCGGACGGACUGCGGAAAUUUGUUGCCGCUGCUUAGAUGCCCGUAAAAUUGCA ....((.((((....(((.(((.((...))))).)))....)))))).(.((((((((((.(((...((((.(((...((((....))))..)))))))...))).)))).)))))).). ( -33.00) >consensus AAACUGCUGACCAAAUCAAUUUCGCAAUGCAAAAUGAAAAUGUUACAAGGGAUUUUCGGGGAUCGAUGCGGACGGACUGCGGAAAUUUGUUGCCGCUGCUUAGAUGCCCGUAAAAUUGCA .....((...((....((.(((((..........))))).))......))((((((((((.(((...((((.(((...((((....))))..)))))))...))).)))).)))))))). (-31.43 = -31.43 + 0.00)

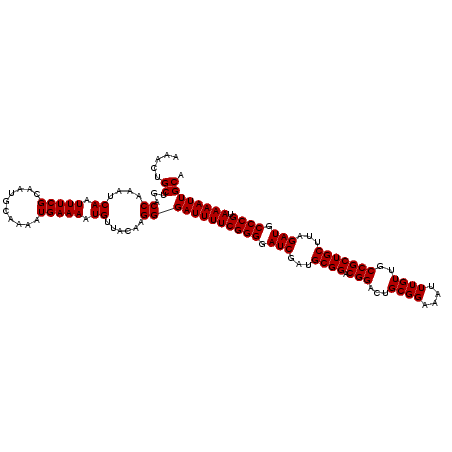

| Location | 18,701,675 – 18,701,785 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.24 |
| Mean single sequence MFE | -35.93 |
| Consensus MFE | -34.42 |
| Energy contribution | -34.42 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.96 |
| SVM decision value | 2.52 |
| SVM RNA-class probability | 0.994856 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18701675 110 + 23771897 UGUUACAAGGGAUUUUCGGGGAUCGAUGCGGACGGACUGCGGAAAUUUGUUGCCGCUGCUUAGAUGCCCGUAAAAUUGCAUGCGGAAAAGCAGCAC----------AAACAUUACGUACC ((((......((((((((((.(((...((((.(((...((((....))))..)))))))...))).)))).))))))((.(((......)))))..----------.))))......... ( -34.60) >DroSec_CAF1 31099 110 + 1 UGUUACAAGGGAUUUUCGGGGAUCGAUGCGGACGGACUGCGGAAAUUUGUUGCCGCUGCUUAGAUGCCCGUAAAAUUGCAUGCGGAAAAGCAGCAC----------AAACAUUACGUACC ((((......((((((((((.(((...((((.(((...((((....))))..)))))))...))).)))).))))))((.(((......)))))..----------.))))......... ( -34.60) >DroAna_CAF1 30196 120 + 1 UGUUAGAGGGGAUUUUCGGGGAUCGAUGCGGACGGACUGCGGAAAUUUGUUACCGCUGCUUAGAUGCCCGUAAAAUUGCAUGCGGAAAAGCAGCAGCAGCCCCACAAAACAUUAGAUACG .......(((((((((((((.(((...((((.(((...((((....))))..)))))))...))).)))).)))))(((.(((......))))))....))))................. ( -39.80) >DroSim_CAF1 32273 110 + 1 UGUUACAAGGGAUUUUCGGGGAUCGAUGCGGACGGACUGCGGAAAUUUGUUGCCGCUGCUUAGAUGCCCGUAAAAUUGCAUGCGGAAAAGCAGCAC----------AAACAUUACGUACC ((((......((((((((((.(((...((((.(((...((((....))))..)))))))...))).)))).))))))((.(((......)))))..----------.))))......... ( -34.60) >DroEre_CAF1 31665 110 + 1 UGUUACAGGGGAUUUUCGGGGAUCGAUGCGGACGGAAUGCGGAAAUUUGUUGCCGCUGCUUAGAUGCCCGUAAAAUUGCAUGCGGAAAAGCAGCAC----------AAACAUUACGUAAC .(((((....((((((((((.(((...((((.(((...((((....))))..)))))))...))).)))).))))))((.(((......)))))..----------.........))))) ( -37.40) >DroYak_CAF1 32538 110 + 1 UGUUACAAGGGAUUUUCGGGGAUCGAUGCGGACGGACUGCGGAAAUUUGUUGCCGCUGCUUAGAUGCCCGUAAAAUUGCAUGCGGAAAAGCAGCAC----------AAACAUUACGUACC ((((......((((((((((.(((...((((.(((...((((....))))..)))))))...))).)))).))))))((.(((......)))))..----------.))))......... ( -34.60) >consensus UGUUACAAGGGAUUUUCGGGGAUCGAUGCGGACGGACUGCGGAAAUUUGUUGCCGCUGCUUAGAUGCCCGUAAAAUUGCAUGCGGAAAAGCAGCAC__________AAACAUUACGUACC ((((......((((((((((.(((...((((.(((...((((....))))..)))))))...))).)))).))))))((.(((......))))).............))))......... (-34.42 = -34.42 + -0.00)


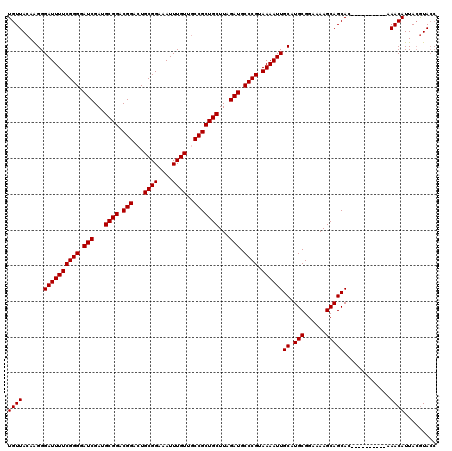
| Location | 18,701,755 – 18,701,850 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 90.84 |
| Mean single sequence MFE | -31.40 |
| Consensus MFE | -22.04 |
| Energy contribution | -22.12 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.597082 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18701755 95 + 23771897 UGCGGAAAAGCAGCACAAACAUUACGUACCAGGAUUGCGAGCGGAAAAA-CUACGACUACG----GUUUUUCCAAAAGGUGCAGC---GUUGUUGUGCUGCAA .........(((((((((.((....(((((......(....)(((((((-((........)----))))))))....)))))...---..))))))))))).. ( -32.90) >DroSec_CAF1 31179 99 + 1 UGCGGAAAAGCAGCACAAACAUUACGUACCCGGAUUGCGAGCGGAAAAA-CUACGACUACGACGGGGUUUUCCAAAAGGUGCAGC---GUUGUUGUGCUGCAA .........(((((((((.((..((((.((((.....((((((......-...)).)).)).))))((...((....)).)).))---))))))))))))).. ( -31.40) >DroSim_CAF1 32353 99 + 1 UGCGGAAAAGCAGCACAAACAUUACGUACCCGGAUUGCGAGCGGAAAAA-CUGCGACUACGACGGGGUUUUCCGAAAGGUGCAGC---GUUGUUGCGCUGCAA (((((....((((((........((((.((((........((((.....-))))........))))((...((....)).)).))---)))))))).))))). ( -33.69) >DroEre_CAF1 31745 95 + 1 UGCGGAAAAGCAGCACAAACAUUACGUAACCGGAUUGCGAGCGGGAAAACCUACGACUACG----GGUUUUCCA-AAGGUGCAGC---GUUGUUGUGCGGCAA .........((.((((((.((..((((.(((....((....))(((((((((........)----)))))))).-..)))...))---)))))))))).)).. ( -30.30) >DroYak_CAF1 32618 97 + 1 UGCGGAAAAGCAGCACAAACAUUACGUACCAGGAUUGCGAGCGGAAAAA-CUACGACUACG----AAUUUUCCA-AAGGUGCAGCAGUGUUGUUGUGUUGCAA .........((((((((((((((.((((((.(((...((((((......-...)).)).))----.....))).-..))))).).)))))..))))))))).. ( -28.70) >consensus UGCGGAAAAGCAGCACAAACAUUACGUACCCGGAUUGCGAGCGGAAAAA_CUACGACUACG____GGUUUUCCAAAAGGUGCAGC___GUUGUUGUGCUGCAA .........(((((((((.((((.((((.......))))...((((((.....((....))......))))))....))))((((...))))))))))))).. (-22.04 = -22.12 + 0.08)


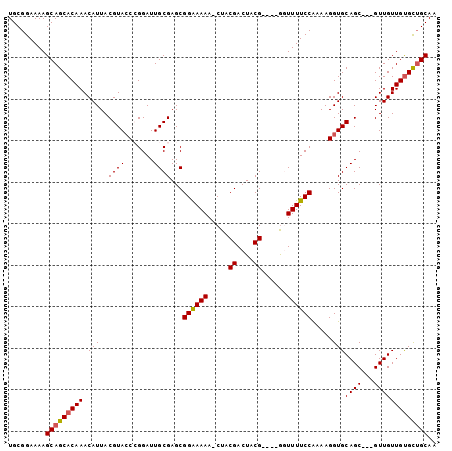
| Location | 18,701,850 – 18,701,943 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 93.18 |
| Mean single sequence MFE | -19.08 |
| Consensus MFE | -17.02 |
| Energy contribution | -17.22 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.89 |
| SVM decision value | 2.29 |
| SVM RNA-class probability | 0.991885 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18701850 93 - 23771897 AAUCUCCCCCUUUUUGUGGCACGUACACGUGCAUCCAUCCACACCCACGUCCAUUUCAAUGGCGUCUAAUAUGCGCUCAAAUGCUGGUACAAU .............((((((((((....)))))..(((.........(((.((((....))))))).........((......)))))))))). ( -19.00) >DroSec_CAF1 31278 92 - 1 AAUCUCCCC-AUUUUGUGGCACGUACACGUGCACCCAUCCACACCCACGUCCAUUUCAAUGGCCUCUAAUAUGCGCUCAAACGCUGGCACAAU .........-...((((((((((....)))))..(((...........(.((((....)))).)........(((......))))))))))). ( -19.30) >DroSim_CAF1 32452 92 - 1 AAUCUCCCC-AUUUUGUGGCACGUACACGUGCACCCAUCCACACCCACGUCCAUUUCAAUGGCCUCUAAUAUGCGCUCAAACGCUGGCACAAU .........-...((((((((((....)))))..(((...........(.((((....)))).)........(((......))))))))))). ( -19.30) >DroEre_CAF1 31840 92 - 1 AAUCUGCCC-AUUUUGUGGCACGUACACGUGCACCUUGCCACACCCACGUCCAAUUCAAUGGCGUCUAAUAUGCGCUCAAACGCUGGUACAAU ....((((.-....(((((((.(((....)))....)))))))...(((.(((......)))))).......(((......))).)))).... ( -20.60) >DroYak_CAF1 32715 92 - 1 AAUCUCCCC-AUUUUGUGGCACGUACACAUGCACCCCGCCACACCCACGUCCAAUUCAAUGGCGUCUAAUAUGCGCACAAACGCUGGCACAAU .........-...((((.(((.(....).))).....((((.....(((.(((......)))))).......(((......))))))))))). ( -17.20) >consensus AAUCUCCCC_AUUUUGUGGCACGUACACGUGCACCCAUCCACACCCACGUCCAUUUCAAUGGCGUCUAAUAUGCGCUCAAACGCUGGCACAAU .............((((((((((....)))))......(((.....(((.(((......)))))).......(((......))))))))))). (-17.02 = -17.22 + 0.20)


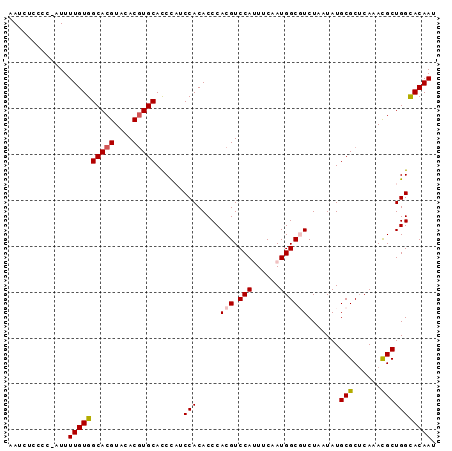
| Location | 18,701,910 – 18,702,015 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 88.32 |
| Mean single sequence MFE | -38.20 |
| Consensus MFE | -26.20 |
| Energy contribution | -26.60 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.76 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.861889 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18701910 105 + 23771897 UGCACGUGUACGUGCCACAAAAAGGGGGAGAUUCACCACCAGA-AGCACGAAUUUUGCAGCAGAUGCAGCCGAGAUCCC-GAAAAGGGAUCAACAGGUGGGGAAUCC- .(((((....)))))..............(((((.(((((...-..........(((((.....)))))....((((((-.....))))))....))))).))))).- ( -37.70) >DroSec_CAF1 31338 104 + 1 UGCACGUGUACGUGCCACAAAAU-GGGGAGAUUC--CGCCAGAAAGCACGAAUUUCGCAGCAGAUGCAGCCCAGAUCCC-GAAAAGGGAUCAACAGGUGCGGAAUCCG .((((.(((.............(-((((((((((--.((......))..)))))))(((.....)))..))))((((((-.....)))))).))).))))........ ( -38.00) >DroSim_CAF1 32512 104 + 1 UGCACGUGUACGUGCCACAAAAU-GGGGAGAUUC--CGCCAGAAAGCACGAAUUUCGCAGCAGAUGCAGCCCAGAUCCC-GAAAAGGGAUCAACAGGUGCGGAAUCCG .((((.(((.............(-((((((((((--.((......))..)))))))(((.....)))..))))((((((-.....)))))).))).))))........ ( -38.00) >DroEre_CAF1 31900 103 + 1 UGCACGUGUACGUGCCACAAAAU-GGGCAGAUUC--CGCCAGC-AGCACGAAAUUUGCAGCAGAUGCAGCGCAGAUCCC-AAAAAGGGAUCAACAGGUUGGGCAUCCG .(((((....)))))........-.(((......--.))).((-.(((.......))).)).(((((...((.((((((-.....)))))).....))...))))).. ( -31.60) >DroYak_CAF1 32775 104 + 1 UGCAUGUGUACGUGCCACAAAAU-GGGGAGAUUC--CACCAGA-AGCUGCAAAUUUGCAGCAGCUGCAGCGCGGAUCCCCAAAAGGGGAUCAACAGGUGGGGCAGCCC .(((((....)))))........-.(((...(((--((((...-.((((((....)))))).((....))(..(((((((....)))))))..).)))))))...))) ( -45.70) >consensus UGCACGUGUACGUGCCACAAAAU_GGGGAGAUUC__CGCCAGA_AGCACGAAUUUUGCAGCAGAUGCAGCCCAGAUCCC_GAAAAGGGAUCAACAGGUGGGGAAUCCG .(((((....)))))..............(((((..((((................((.((....)).))...((((((......))))))....))))..))))).. (-26.20 = -26.60 + 0.40)


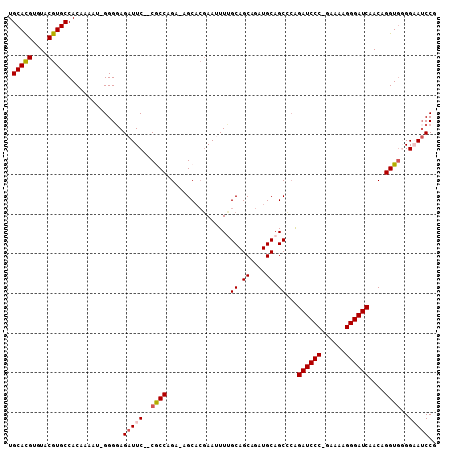
| Location | 18,701,910 – 18,702,015 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 88.32 |
| Mean single sequence MFE | -39.36 |
| Consensus MFE | -24.78 |
| Energy contribution | -25.58 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.86 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.621824 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18701910 105 - 23771897 -GGAUUCCCCACCUGUUGAUCCCUUUUC-GGGAUCUCGGCUGCAUCUGCUGCAAAAUUCGUGCU-UCUGGUGGUGAAUCUCCCCCUUUUUGUGGCACGUACACGUGCA -((((((.(((((....((((((.....-))))))...((.((....)).))............-...))))).)))))).............(((((....))))). ( -37.40) >DroSec_CAF1 31338 104 - 1 CGGAUUCCGCACCUGUUGAUCCCUUUUC-GGGAUCUGGGCUGCAUCUGCUGCGAAAUUCGUGCUUUCUGGCG--GAAUCUCCCC-AUUUUGUGGCACGUACACGUGCA .(((((((((.((....((((((.....-)))))).))((.((....)).)).................)))--))))))....-........(((((....))))). ( -38.90) >DroSim_CAF1 32512 104 - 1 CGGAUUCCGCACCUGUUGAUCCCUUUUC-GGGAUCUGGGCUGCAUCUGCUGCGAAAUUCGUGCUUUCUGGCG--GAAUCUCCCC-AUUUUGUGGCACGUACACGUGCA .(((((((((.((....((((((.....-)))))).))((.((....)).)).................)))--))))))....-........(((((....))))). ( -38.90) >DroEre_CAF1 31900 103 - 1 CGGAUGCCCAACCUGUUGAUCCCUUUUU-GGGAUCUGCGCUGCAUCUGCUGCAAAUUUCGUGCU-GCUGGCG--GAAUCUGCCC-AUUUUGUGGCACGUACACGUGCA (((((((......(((.((((((.....-)))))).)))..)))))))..(((.....(((((.-.(.((((--(...))))).-.....)..)))))......))). ( -39.80) >DroYak_CAF1 32775 104 - 1 GGGCUGCCCCACCUGUUGAUCCCCUUUUGGGGAUCCGCGCUGCAGCUGCUGCAAAUUUGCAGCU-UCUGGUG--GAAUCUCCCC-AUUUUGUGGCACGUACACAUGCA (((..(..(((((.((.(((((((....))))))).)).....((..((((((....)))))).-.))))))--)...)..)))-....((((.......)))).... ( -41.80) >consensus CGGAUUCCCCACCUGUUGAUCCCUUUUC_GGGAUCUGGGCUGCAUCUGCUGCAAAAUUCGUGCU_UCUGGCG__GAAUCUCCCC_AUUUUGUGGCACGUACACGUGCA .((((((....((....((((((......))))))...((.((....)).))................))....)))))).............(((((....))))). (-24.78 = -25.58 + 0.80)


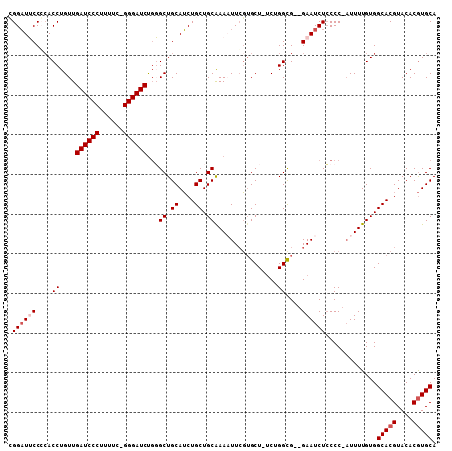
| Location | 18,701,943 – 18,702,033 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 75.84 |
| Mean single sequence MFE | -43.27 |
| Consensus MFE | -22.95 |
| Energy contribution | -23.32 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.17 |
| Mean z-score | -3.53 |
| Structure conservation index | 0.53 |
| SVM decision value | 3.60 |
| SVM RNA-class probability | 0.999433 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18701943 90 + 23771897 CACCACCAGA-AGCACGAAUUUUGCAGCAGAUGCAGCCGAGAUCCC-GAAAAGGGAUCAACAGGUGGGGAAUCC--------------AGCUGGCGAGCAU------GUGCA ..........-.(((.......))).(((.((((.(((..((((((-.....))))))...((.(((.....))--------------).)))))..))))------.))). ( -34.50) >DroSim_CAF1 32544 103 + 1 C--CGCCAGAAAGCACGAAUUUCGCAGCAGAUGCAGCCCAGAUCCC-GAAAAGGGAUCAACAGGUGCGGAAUCCGAGUGGGAGAAUCCAGCUGGCGAGCAU------GUGCA .--((((((...((((.......(((.....)))......((((((-.....)))))).....))))(((.(((.....)))...)))..)))))).((..------..)). ( -40.00) >DroEre_CAF1 31932 108 + 1 C--CGCCAGC-AGCACGAAAUUUGCAGCAGAUGCAGCGCAGAUCCC-AAAAAGGGAUCAACAGGUUGGGCAUCCGAGUGGGAGAAUCCAGCUGCCGGGUUUUCCCGGCUGCA .--.....((-.(((.......))).))...((((((((.((((((-.....))))))....(((((((..(((.....)))...)))))))))((((....)))))))))) ( -44.10) >DroYak_CAF1 32807 109 + 1 C--CACCAGA-AGCUGCAAAUUUGCAGCAGCUGCAGCGCGGAUCCCCAAAAGGGGAUCAACAGGUGGGGCAGCCCAGUGGGAAAAUCCAGCUAGCGAGCUUUCCCGGCUGCA (--((((...-.((((((....)))))).((....))(..(((((((....)))))))..).))))).((((((....(((((.....((((....))))))))))))))). ( -54.50) >consensus C__CACCAGA_AGCACGAAAUUUGCAGCAGAUGCAGCCCAGAUCCC_AAAAAGGGAUCAACAGGUGGGGAAUCCGAGUGGGAGAAUCCAGCUGGCGAGCAU______CUGCA ............((((..((((((((((.(((((.(((..((((((......))))))....)))...)))))......((.....)).))).))))).))......)))). (-22.95 = -23.32 + 0.37)



| Location | 18,701,943 – 18,702,033 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 75.84 |
| Mean single sequence MFE | -40.15 |
| Consensus MFE | -20.74 |
| Energy contribution | -20.55 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.52 |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.902853 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18701943 90 - 23771897 UGCAC------AUGCUCGCCAGCU--------------GGAUUCCCCACCUGUUGAUCCCUUUUC-GGGAUCUCGGCUGCAUCUGCUGCAAAAUUCGUGCU-UCUGGUGGUG .(((.------((((..(((((.(--------------((.....))).))...((((((.....-))))))..))).)))).))).(((.......))).-.......... ( -27.40) >DroSim_CAF1 32544 103 - 1 UGCAC------AUGCUCGCCAGCUGGAUUCUCCCACUCGGAUUCCGCACCUGUUGAUCCCUUUUC-GGGAUCUGGGCUGCAUCUGCUGCGAAAUUCGUGCUUUCUGGCG--G .((..------..)).((((((..(((...(((.....))).)))((((((...((((((.....-)))))).))).)))....((.(((.....)))))...))))))--. ( -36.50) >DroEre_CAF1 31932 108 - 1 UGCAGCCGGGAAAACCCGGCAGCUGGAUUCUCCCACUCGGAUGCCCAACCUGUUGAUCCCUUUUU-GGGAUCUGCGCUGCAUCUGCUGCAAAUUUCGUGCU-GCUGGCG--G .(((((((((....))))((((((((......)))...((((((......(((.((((((.....-)))))).)))..))))))))))).........)))-)).....--. ( -43.70) >DroYak_CAF1 32807 109 - 1 UGCAGCCGGGAAAGCUCGCUAGCUGGAUUUUCCCACUGGGCUGCCCCACCUGUUGAUCCCCUUUUGGGGAUCCGCGCUGCAGCUGCUGCAAAUUUGCAGCU-UCUGGUG--G .(((((((((((((..((.....))..)))))))....)))))).(((((.((.(((((((....))))))).)).....((..((((((....)))))).-.))))))--) ( -53.00) >consensus UGCAC______AAGCUCGCCAGCUGGAUUCUCCCACUCGGAUGCCCCACCUGUUGAUCCCUUUUC_GGGAUCUGCGCUGCAUCUGCUGCAAAAUUCGUGCU_UCUGGCG__G ................((((((................................((((((......))))))...((.((....)).))..............))))))... (-20.74 = -20.55 + -0.19)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:33:33 2006