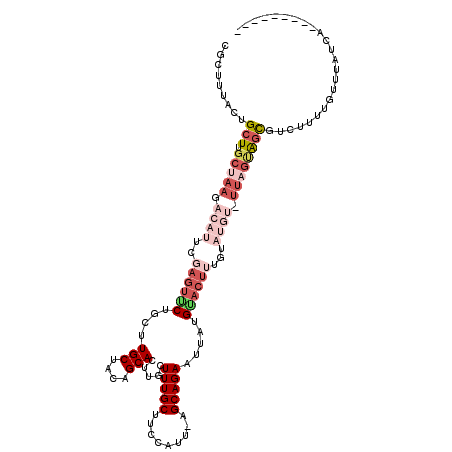
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 18,688,794 – 18,688,980 |
| Length | 186 |
| Max. P | 0.962828 |

| Location | 18,688,794 – 18,688,914 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.24 |
| Mean single sequence MFE | -31.82 |
| Consensus MFE | -14.56 |
| Energy contribution | -16.80 |
| Covariance contribution | 2.24 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.46 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.626589 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18688794 120 + 23771897 AUAUAUUGAGCUCUUUAAGAUCUGAAAGAUAAUUACGCAUUCAGAUUCUGGGAACCCCUAUGGAUAAUCUUUAAAGCAGUCGCUUUACUGCUUGCUAAGACAUUCGUGUUCUGCUUGCUA .......((((....((..(((((((.(.........).)))))))..))(((((....((((((..((((..(((((((......)))))))...)))).))))))))))))))).... ( -28.20) >DroSec_CAF1 18155 119 + 1 AGAUAUUGAGCUCUUUAAGAUCUGGAAGAUAAUUAUGCAUGCAGAUUCUGGGAAC-CCUAUGGACAAUAUUUAAAGCAGUCGCUUUACUGCUUGCUAAGACAUUCCAGUUCUGCUUGCUA ........(((......(((.((((((.....(((.(((.((((.(((.((....-))...))).......((((((....)))))))))).))))))....)))))).)))....))). ( -32.50) >DroSim_CAF1 18151 120 + 1 AGAUAUUGAGCUCUUUAAGAUCUGGAAGAUAAUUACGCAUGCAGAUUCUGGGAACCCCUAUGGACACUAUUUAAAGCAGUCGCUUUACUGCUUGCUAAGACAUUCCAAUUCUGCAUGCUA ...........(((((........))))).......(((((((((...((((....))))((((.....((..(((((((......)))))))...)).....))))..))))))))).. ( -32.30) >DroEre_CAF1 18173 107 + 1 AGAUAGCGAGCACUUCAAGAACGGUAAGCUGAUUGCGCAUGCAGAUUCUG----------UGCACAAUCUCUAAAGCAGUGGC---ACUGCUUGCUAAGGCACUCGAGUGCUGCUUGCUA ...((((((((...........(((((((.(((((.((((.........)----------))).)))))......((....))---...)))))))..(((((....))))))))))))) ( -37.80) >DroYak_CAF1 18701 107 + 1 AGGUAAUAAGCACUUAAAGAUCUGAAAGAUGAUUUCGCAUGCAGAUUCUU----------UGCAGAAUCUGUUAAGCAUUCGC---ACUGCUUGCUAAGGCACUCGAGCACUGCUUGCUG ........((((......((...((((.....))))((..(((((((((.----------...)))))))))...))..))((---(.(((((((....)))....)))).))).)))). ( -28.30) >consensus AGAUAUUGAGCUCUUUAAGAUCUGAAAGAUAAUUACGCAUGCAGAUUCUGGGAAC_CCUAUGGACAAUCUUUAAAGCAGUCGCUUUACUGCUUGCUAAGACAUUCGAGUUCUGCUUGCUA ....................................(((.((((...((((((....................(((((((......)))))))(......).))))))..)))).))).. (-14.56 = -16.80 + 2.24)


| Location | 18,688,794 – 18,688,914 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.24 |
| Mean single sequence MFE | -30.88 |
| Consensus MFE | -15.94 |
| Energy contribution | -16.50 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.739192 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18688794 120 - 23771897 UAGCAAGCAGAACACGAAUGUCUUAGCAAGCAGUAAAGCGACUGCUUUAAAGAUUAUCCAUAGGGGUUCCCAGAAUCUGAAUGCGUAAUUAUCUUUCAGAUCUUAAAGAGCUCAAUAUAU ..(((..((((....((..(((((...(((((((......)))))))..)))))..))....(((...)))....))))..))).......(((((........)))))........... ( -25.10) >DroSec_CAF1 18155 119 - 1 UAGCAAGCAGAACUGGAAUGUCUUAGCAAGCAGUAAAGCGACUGCUUUAAAUAUUGUCCAUAGG-GUUCCCAGAAUCUGCAUGCAUAAUUAUCUUCCAGAUCUUAAAGAGCUCAAUAUCU ..(((.(((((..((((..........(((((((......))))))).........))))..((-....))....))))).)))..............((.(((...))).))....... ( -27.21) >DroSim_CAF1 18151 120 - 1 UAGCAUGCAGAAUUGGAAUGUCUUAGCAAGCAGUAAAGCGACUGCUUUAAAUAGUGUCCAUAGGGGUUCCCAGAAUCUGCAUGCGUAAUUAUCUUCCAGAUCUUAAAGAGCUCAAUAUCU ..(((((((((.(((((((.(((..(((.....((((((....)))))).....)))....))).)))))..)).)))))))))..............((.(((...))).))....... ( -32.60) >DroEre_CAF1 18173 107 - 1 UAGCAAGCAGCACUCGAGUGCCUUAGCAAGCAGU---GCCACUGCUUUAGAGAUUGUGCA----------CAGAAUCUGCAUGCGCAAUCAGCUUACCGUUCUUGAAGUGCUCGCUAUCU .....(((((((((((((.((...(((((((((.---....))))))....(((((((((----------(((...)))..))))))))).)))....)).)))).)))))).))).... ( -40.90) >DroYak_CAF1 18701 107 - 1 CAGCAAGCAGUGCUCGAGUGCCUUAGCAAGCAGU---GCGAAUGCUUAACAGAUUCUGCA----------AAGAAUCUGCAUGCGAAAUCAUCUUUCAGAUCUUUAAGUGCUUAUUACCU .(((..(((.((((....(((....))))))).)---))....((((((((((((((...----------.)))))))).....((((.....))))......)))))))))........ ( -28.60) >consensus UAGCAAGCAGAACUCGAAUGUCUUAGCAAGCAGUAAAGCGACUGCUUUAAAGAUUGUCCAUAGG_GUUCCCAGAAUCUGCAUGCGUAAUUAUCUUCCAGAUCUUAAAGAGCUCAAUAUCU ..(((.(((((........((....))(((((((......)))))))............................))))).))).................................... (-15.94 = -16.50 + 0.56)


| Location | 18,688,834 – 18,688,949 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.24 |
| Mean single sequence MFE | -32.12 |
| Consensus MFE | -15.96 |
| Energy contribution | -15.92 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.50 |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.962828 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18688834 115 + 23771897 UCAGAUUCUGGGAACCCCUAUGGAUAAUCUUUAAAGCAGUCGCUUUACUGCUUGCUAAGACAUUCGUGUUCUGCUUGCUACAGCA-----UUUGCUUCGAUUAAGCAGAAUUAUGUACUU ....((((((((....)))).))))...(((..(((((((......)))))))...)))((((....((((((((((.(..(((.-----...)))...).)))))))))).)))).... ( -31.20) >DroSec_CAF1 18195 118 + 1 GCAGAUUCUGGGAAC-CCUAUGGACAAUAUUUAAAGCAGUCGCUUUACUGCUUGCUAAGACAUUCCAGUUCUGCUUGCUACGGCACUUUCUUUGCUUCCAUU-AGCAGAAUUAUGAACUU (((((..((((((.(-(....))..........(((((((......))))))).........)))))).))))).(((....))).....((((((......-))))))........... ( -33.20) >DroSim_CAF1 18191 119 + 1 GCAGAUUCUGGGAACCCCUAUGGACACUAUUUAAAGCAGUCGCUUUACUGCUUGCUAAGACAUUCCAAUUCUGCAUGCUACGGCACUUUCUUUGCUUCCAUU-AGCAGAAUUAUGAACUU (((((...((((....))))((((.....((..(((((((......)))))))...)).....))))..))))).(((....))).....((((((......-))))))........... ( -31.70) >DroEre_CAF1 18213 106 + 1 GCAGAUUCUG----------UGCACAAUCUCUAAAGCAGUGGC---ACUGCUUGCUAAGGCACUCGAGUGCUGCUUGCUACAGCACUAGCUUUGCUUCCAUU-UGCAGAAUUAUGUACUU ((((((((((----------(............((((((....---.))))))((.(((((.....(((((((.......))))))).))))))).......-.)))))))).))).... ( -32.70) >DroYak_CAF1 18741 106 + 1 GCAGAUUCUU----------UGCAGAAUCUGUUAAGCAUUCGC---ACUGCUUGCUAAGGCACUCGAGCACUGCUUGCUGCAGCACAUGCUUUGCUUUCAUU-UGCAGAAUUAUGUACUU (((((((((.----------...)))))))))...((((....---.((((..(..((((((...(((((((((.....))))....)))))))))))..).-.))))....)))).... ( -31.80) >consensus GCAGAUUCUGGGAAC_CCUAUGGACAAUCUUUAAAGCAGUCGCUUUACUGCUUGCUAAGACAUUCGAGUUCUGCUUGCUACAGCACUUGCUUUGCUUCCAUU_AGCAGAAUUAUGUACUU ...(((((((.......................(((((((......)))))))((..((((......)).))....))...((((.......)))).........)))))))........ (-15.96 = -15.92 + -0.04)


| Location | 18,688,874 – 18,688,980 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.86 |
| Mean single sequence MFE | -26.72 |
| Consensus MFE | -11.35 |
| Energy contribution | -13.31 |
| Covariance contribution | 1.96 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.42 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.665766 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18688874 106 + 23771897 CGCUUUACUGCUUGCUAAGACAUUCGUGUUCUGCUUGCUACAGCA-----UUUGCUUCGAUUAAGCAGAAUUAUGUACUUUGUAUGUAUUAGUAGCGUCUUUUGUUUAUCA--------- (((((((.(((.(((.(((((((....((((((((((.(..(((.-----...)))...).)))))))))).)))).))).))).))).))).))))..............--------- ( -28.40) >DroSec_CAF1 18234 110 + 1 CGCUUUACUGCUUGCUAAGACAUUCCAGUUCUGCUUGCUACGGCACUUUCUUUGCUUCCAUU-AGCAGAAUUAUGAACUUUGAAUGUGUUUGUAGCGUCUUUUGUUUAUCA--------- .........(((.((.((.((((((.(((((....(((....))).....((((((......-)))))).....)))))..)))))).)).)))))...............--------- ( -26.70) >DroSim_CAF1 18231 110 + 1 CGCUUUACUGCUUGCUAAGACAUUCCAAUUCUGCAUGCUACGGCACUUUCUUUGCUUCCAUU-AGCAGAAUUAUGAACUUUGUAUGUGUUAGUAGCGUCUGUUGUUUAUCA--------- .........(((.(((((.((((..(((.....(((((....))......((((((......-))))))...)))....))).)))).))))))))...............--------- ( -23.80) >DroEre_CAF1 18243 107 + 1 GGC---ACUGCUUGCUAAGGCACUCGAGUGCUGCUUGCUACAGCACUAGCUUUGCUUCCAUU-UGCAGAAUUAUGUACUUGGUA----UUAGCAGCGUAUUUUGUUUAUCCCUAA----- .((---.(((((((((((((((....(((((((.......)))))))...(((((.......-.)))))....))).)))))))----..))))).)).................----- ( -31.00) >DroYak_CAF1 18771 112 + 1 CGC---ACUGCUUGCUAAGGCACUCGAGCACUGCUUGCUGCAGCACAUGCUUUGCUUUCAUU-UGCAGAAUUAUGUACUUUGUA----AAUAAGUUGUAUUUUGUUGAUCCUUGUUGAUC .((---(.(((((((....)))....)))).)))......(((((.((((...((((..(((-((((((.........))))))----))))))).))))..)))))............. ( -23.70) >consensus CGCUUUACUGCUUGCUAAGACAUUCGAGUUCUGCUUGCUACAGCACUUGCUUUGCUUCCAUU_AGCAGAAUUAUGUACUUUGUAUGU_UUAGUAGCGUCUUUUGUUUAUCA_________ ((((........(((.(((((((...((((((((.......((((.......))))........)))))))))))).))).))).........))))....................... (-11.35 = -13.31 + 1.96)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:33:13 2006