
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 18,686,863 – 18,687,009 |
| Length | 146 |
| Max. P | 0.994614 |

| Location | 18,686,863 – 18,686,969 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 85.50 |
| Mean single sequence MFE | -22.90 |
| Consensus MFE | -18.81 |
| Energy contribution | -19.97 |
| Covariance contribution | 1.16 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.866002 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18686863 106 - 23771897 CCAUUAACUGCAUUUCAGUGAAAUGUCAACUGGAUUUUUCUCGUACCAGUUUUCCGCUUGGCUCGUCGCUGAUGUGUUUCAGUUGC-UUUAUAUGUAUUUAUUUUUG ....(((((((((.(((((((...(((((.((((.....((......))...)))).)))))...))))))).)))...)))))).-.................... ( -25.70) >DroSec_CAF1 16142 107 - 1 CCAUUAACUGCAUUUCAGUGAAAUGUCAACUGGAUUUUUCUCGUACCAGUUUUCCGCUUGGCUCGUCGCUGAUGUGUUUCAGUUGCCUUUAUCUGUAUUUAUUUUUG ....(((((((((.(((((((...(((((.((((.....((......))...)))).)))))...))))))).)))...))))))...................... ( -25.70) >DroAna_CAF1 15531 97 - 1 CCAUUAACUGCAUUUCAGUAAAAUGUCAACUGGAUU-UUCUUGUACCCGUUAUCCACUUGGUUCGCCACUGAUUU--------UAC-UAUCCAUGUGUGUGCUCUUA .........((((.(((((......((((.(((((.-..(........)..))))).))))......)))))...--------(((-.......))).))))..... ( -13.80) >DroSim_CAF1 16168 107 - 1 CCAUUAACUGCAUUUCAGUGAAAUGUCAACUGGAUUUUUCUCGUACCAGUUUUCCGCUUGGCUCGUCGCUGAUGUGUUUCAGUUGCCUUUAUCUGUAUUUAUUUUUG ....(((((((((.(((((((...(((((.((((.....((......))...)))).)))))...))))))).)))...))))))...................... ( -25.70) >DroEre_CAF1 16109 106 - 1 CCAUUAACAGCAUUUCAGUGAAAUGUCAACUGGAUUUUUCUUGUGCCAGUUUUCCGCUUGGCUCGUCGCUGAUGUGUUUCUUUUU-CUGUAGCUGUAUUUGUUUUUG ......(((((...(((((((...(((((.((((.....((......))...)))).)))))...))))))).(.....).....-.....)))))........... ( -23.60) >consensus CCAUUAACUGCAUUUCAGUGAAAUGUCAACUGGAUUUUUCUCGUACCAGUUUUCCGCUUGGCUCGUCGCUGAUGUGUUUCAGUUGCCUUUAUCUGUAUUUAUUUUUG ....(((((((((.(((((((...(((((.((((..................)))).)))))...))))))).)))...))))))...................... (-18.81 = -19.97 + 1.16)



| Location | 18,686,889 – 18,687,009 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.56 |
| Mean single sequence MFE | -31.42 |
| Consensus MFE | -26.96 |
| Energy contribution | -27.43 |
| Covariance contribution | 0.47 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.812826 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18686889 120 + 23771897 GAAACACAUCAGCGACGAGCCAAGCGGAAAACUGGUACGAGAAAAAUCCAGUUGACAUUUCACUGAAAUGCAGUUAAUGGGAAUUCUCCUGGCCAUCUCUGAAGAUAACGAUUGCUAAUG ......(((.(((((((.......((((....((((..(((((...((((.(((.((((((...)))))))))....))))..)))))...))))..)))).......)).))))).))) ( -28.14) >DroSec_CAF1 16169 120 + 1 GAAACACAUCAGCGACGAGCCAAGCGGAAAACUGGUACGAGAAAAAUCCAGUUGACAUUUCACUGAAAUGCAGUUAAUGGGAAUUCGCCUGGCCAUCUCUGAAGAUAACGAUUGCUAAUG ......(((.(((((((.((((.(((((.((((((............))))))..((((..((((.....)))).))))....))))).)))).((((....))))..)).))))).))) ( -32.70) >DroAna_CAF1 15554 114 + 1 -----AAAUCAGUGGCGAACCAAGUGGAUAACGGGUACAAGAA-AAUCCAGUUGACAUUUUACUGAAAUGCAGUUAAUGGGAAUUCGCUUGGCUAUCUCUGAGGAUAACGAUUGCUAAUG -----.......((((((.((((((((((..(((((.......-.))))......((((..((((.....)))).)))))..)))))))))).(((((....)))))....))))))... ( -30.10) >DroSim_CAF1 16195 120 + 1 GAAACACAUCAGCGACGAGCCAAGCGGAAAACUGGUACGAGAAAAAUCCAGUUGACAUUUCACUGAAAUGCAGUUAAUGGGAAUUCGCUUGGCCAUCUCUGAAGAUAACGAUUGCUAAUG ......(((.(((((((.((((((((((.((((((............))))))..((((..((((.....)))).))))....)))))))))).((((....))))..)).))))).))) ( -36.80) >DroEre_CAF1 16135 120 + 1 GAAACACAUCAGCGACGAGCCAAGCGGAAAACUGGCACAAGAAAAAUCCAGUUGACAUUUCACUGAAAUGCUGUUAAUGGGAAUUCGCCUGGCCAACUCUGAAGAUAACGAUUGCUAAUG ......(((.(((((((.((((.(((((...((......)).....(((.(((((((((((...))))...))))))).))).))))).))))....((....))...)).))))).))) ( -28.10) >DroYak_CAF1 16622 120 + 1 GAAACACAUCAGCGACGAGCCAAGCGGAAAACUGGUACGAGAAAAAUCCAGUUGACAUUUCACUGAAAUGCAGUUAAUGGGAAUUCGCCUGGCCAUCUCUGAAGAUAACGAUUGCUAAUG ......(((.(((((((.((((.(((((.((((((............))))))..((((..((((.....)))).))))....))))).)))).((((....))))..)).))))).))) ( -32.70) >consensus GAAACACAUCAGCGACGAGCCAAGCGGAAAACUGGUACGAGAAAAAUCCAGUUGACAUUUCACUGAAAUGCAGUUAAUGGGAAUUCGCCUGGCCAUCUCUGAAGAUAACGAUUGCUAAUG ......(((.(((((((.((((.(((((.((((((............))))))..((((..((((.....)))).))))....))))).)))).((((....))))..)).))))).))) (-26.96 = -27.43 + 0.47)

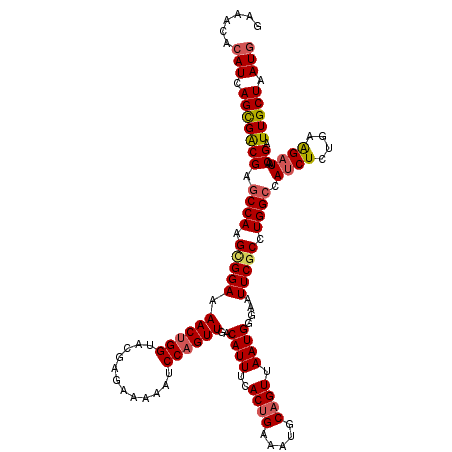

| Location | 18,686,889 – 18,687,009 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.56 |
| Mean single sequence MFE | -33.48 |
| Consensus MFE | -29.86 |
| Energy contribution | -31.00 |
| Covariance contribution | 1.14 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.89 |
| SVM decision value | 2.49 |
| SVM RNA-class probability | 0.994614 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18686889 120 - 23771897 CAUUAGCAAUCGUUAUCUUCAGAGAUGGCCAGGAGAAUUCCCAUUAACUGCAUUUCAGUGAAAUGUCAACUGGAUUUUUCUCGUACCAGUUUUCCGCUUGGCUCGUCGCUGAUGUGUUUC (((((((........((....))((((((((((.(((............(((((((...))))))).((((((............)))))))))..)))))).)))))))))))...... ( -32.50) >DroSec_CAF1 16169 120 - 1 CAUUAGCAAUCGUUAUCUUCAGAGAUGGCCAGGCGAAUUCCCAUUAACUGCAUUUCAGUGAAAUGUCAACUGGAUUUUUCUCGUACCAGUUUUCCGCUUGGCUCGUCGCUGAUGUGUUUC (((((((........((....))((((((((((((..............(((((((...))))))).((((((............))))))...)))))))).)))))))))))...... ( -37.70) >DroAna_CAF1 15554 114 - 1 CAUUAGCAAUCGUUAUCCUCAGAGAUAGCCAAGCGAAUUCCCAUUAACUGCAUUUCAGUAAAAUGUCAACUGGAUU-UUCUUGUACCCGUUAUCCACUUGGUUCGCCACUGAUUU----- .(((((.....((((((......))))))...((((((...((((.((((.....))))..)))).(((.(((((.-..(........)..))))).)))))))))..)))))..----- ( -21.40) >DroSim_CAF1 16195 120 - 1 CAUUAGCAAUCGUUAUCUUCAGAGAUGGCCAAGCGAAUUCCCAUUAACUGCAUUUCAGUGAAAUGUCAACUGGAUUUUUCUCGUACCAGUUUUCCGCUUGGCUCGUCGCUGAUGUGUUUC (((((((........((....))((((((((((((..............(((((((...))))))).((((((............))))))...)))))))).)))))))))))...... ( -38.00) >DroEre_CAF1 16135 120 - 1 CAUUAGCAAUCGUUAUCUUCAGAGUUGGCCAGGCGAAUUCCCAUUAACAGCAUUUCAGUGAAAUGUCAACUGGAUUUUUCUUGUGCCAGUUUUCCGCUUGGCUCGUCGCUGAUGUGUUUC (((((((........((....))...(((((((((..............(((((((...))))))).((((((((.......)).))))))...)))))))))....)))))))...... ( -33.60) >DroYak_CAF1 16622 120 - 1 CAUUAGCAAUCGUUAUCUUCAGAGAUGGCCAGGCGAAUUCCCAUUAACUGCAUUUCAGUGAAAUGUCAACUGGAUUUUUCUCGUACCAGUUUUCCGCUUGGCUCGUCGCUGAUGUGUUUC (((((((........((....))((((((((((((..............(((((((...))))))).((((((............))))))...)))))))).)))))))))))...... ( -37.70) >consensus CAUUAGCAAUCGUUAUCUUCAGAGAUGGCCAGGCGAAUUCCCAUUAACUGCAUUUCAGUGAAAUGUCAACUGGAUUUUUCUCGUACCAGUUUUCCGCUUGGCUCGUCGCUGAUGUGUUUC (((((((........((....))((((((((((((..............(((((((...))))))).((((((............))))))...)))))))).)))))))))))...... (-29.86 = -31.00 + 1.14)

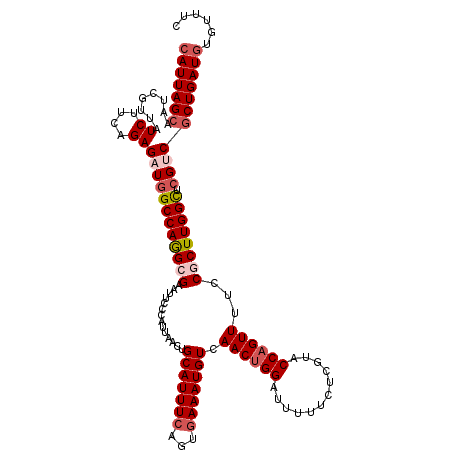
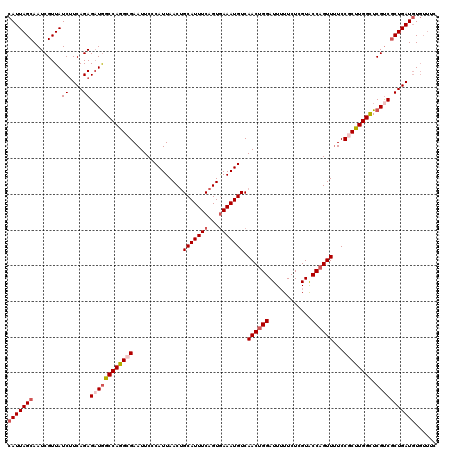
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:33:09 2006