
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 18,670,488 – 18,670,641 |
| Length | 153 |
| Max. P | 0.627397 |

| Location | 18,670,488 – 18,670,602 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.00 |
| Mean single sequence MFE | -28.91 |
| Consensus MFE | -19.58 |
| Energy contribution | -19.97 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.68 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.521888 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18670488 114 + 23771897 ACUUUCUCCCCUUUUAUAGGACUUUUUU--UUUCCC-CCUUGUCCAGGCAGCCGCUUAAUGCAAUCAAAAUGCCUCUGGCUGAGUGCCGGAAUGGCAAUUACUACAGCCACAGUU---AC ..................((((......--......-....)))).(((....((.....))........((((((((((.....))))))..)))).........)))......---.. ( -27.59) >DroSec_CAF1 7754 113 + 1 ACUUUCUCCCCUUUCACAGGACAUUUUU---UUUUG-CCCUAUCCAGGCAGCCGCUUAAUGCAAUCAAAAUGCCGCUGGCCGAGUGCCGGAAUGGCAAUUACUACAGCCACAGUU---AC .................(((.((.....---...))-.))).....(((....((.....))........((((.(((((.....)))))...)))).........)))......---.. ( -24.00) >DroAna_CAF1 7344 110 + 1 AUU-----CCCUUUUGCGUGGCAGAUUU---UUUUGCUUUUAUCCAGGCACCCGCUUAAUGCAAUCAAAAUGCCACUGGUCGAGUGCCGGAAUGGCAAUUGCCGUUGCAAAAGGCA--GC ...-----.(((((((((.((((((...---.))))))....(((.(((((.((((....(((.......)))....)).)).))))))))(((((....))))))))))))))..--.. ( -38.70) >DroSim_CAF1 7695 116 + 1 ACUUUCUCCCCUUUCACAGGACAUUUUUUUUUUUUG-CGUUAUCCAGGCAGCCGCUUAAUGCAAUCAAAAUGCCUCUGGCCGACUGCCGGAAUGGCAAUUACUACAGCCACAGUA---AC .........(((.....))).......((((..(((-(((((....(((....)))))))))))..))))((((((((((.....))))))..)))).(((((........))))---). ( -26.20) >DroEre_CAF1 7666 109 + 1 ACUUUCUCCCCU-UUGCAGGACAUUUUU------UG-CCUUAUCCAGGCAGCCGCCUAAUGCAAUCAAAAUGCCUCUGGCCGAGUGCCGGAAUGGCAAUUACUACUGCCACAGUU---GC ............-..((((........(------((-((...(((.((((.(.(((....(((.......)))....))).)..)))))))..)))))......)))).......---.. ( -29.14) >DroYak_CAF1 7813 113 + 1 ACUUUCUCCCCUUUUGCAGGACAUUUUU------UC-CUUUAUCCUGGCAGCCGCUUAAUGCAAUCAAAAUGCCUCUGGCCGAGUGCCGGAAUGGCAAUUACUACUGCCACAGUUACUGC ...............((((((.......------))-).......((((((..((.....))........((((((((((.....))))))..)))).......)))))).......))) ( -27.80) >consensus ACUUUCUCCCCUUUUACAGGACAUUUUU___UUUUG_CCUUAUCCAGGCAGCCGCUUAAUGCAAUCAAAAUGCCUCUGGCCGAGUGCCGGAAUGGCAAUUACUACAGCCACAGUU___AC .........(((.....)))..........................(((....((.....))........((((((((((.....))))))..)))).........)))........... (-19.58 = -19.97 + 0.39)

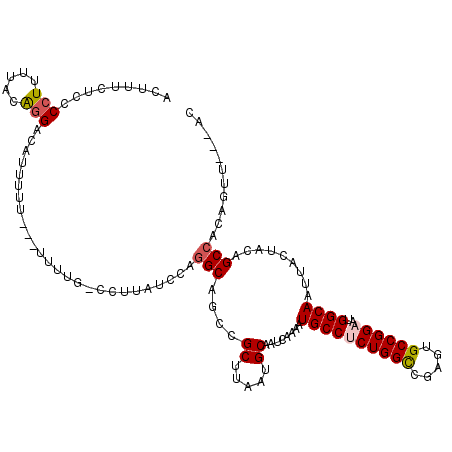

| Location | 18,670,525 – 18,670,641 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.60 |
| Mean single sequence MFE | -33.07 |
| Consensus MFE | -22.15 |
| Energy contribution | -22.75 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.627397 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18670525 116 + 23771897 UGUCCAGGCAGCCGCUUAAUGCAAUCAAAAUGCCUCUGGCUGAGUGCCGGAAUGGCAAUUACUACAGCCACAGUU---ACUG-CACGUCUGCUACCUAUGGCACAGAAUAAAGCCAUUUA ......(((....((.....))..((....((((((((((.....))))))..)))).........((((.((..---...(-((....)))...)).))))...)).....)))..... ( -31.70) >DroSec_CAF1 7790 116 + 1 UAUCCAGGCAGCCGCUUAAUGCAAUCAAAAUGCCGCUGGCCGAGUGCCGGAAUGGCAAUUACUACAGCCACAGUU---ACUG-CACGUCUGCUACCUAUGGCACUGAAUAAAGCCAUUUA ......(((....((.....))..(((...((((.(((((.....)))))...)))).........((((.((..---...(-((....)))...)).))))..))).....)))..... ( -30.40) >DroAna_CAF1 7376 93 + 1 UAUCCAGGCACCCGCUUAAUGCAAUCAAAAUGCCACUGGUCGAGUGCCGGAAUGGCAAUUGCCGUUGCAAAAGGCA--GCUGGAACAGCUGCUAC------------------------- ......(((((.((((....(((.......)))....)).)).)))))(.((((((....)))))).)....((((--((((...))))))))..------------------------- ( -36.00) >DroSim_CAF1 7734 116 + 1 UAUCCAGGCAGCCGCUUAAUGCAAUCAAAAUGCCUCUGGCCGACUGCCGGAAUGGCAAUUACUACAGCCACAGUA---ACUG-CACGUCUGCUACCUAUGGCACUGAAUAAAGCCAUUUA ..(((.((((((.(((....(((.......)))....))).).))))))))(((((..........((((.((..---...(-((....)))...)).))))..........)))))... ( -31.35) >DroEre_CAF1 7698 116 + 1 UAUCCAGGCAGCCGCCUAAUGCAAUCAAAAUGCCUCUGGCCGAGUGCCGGAAUGGCAAUUACUACUGCCACAGUU---GCUG-CACUUCUGUUAGCCAUGGCCCAGAAGAAAGCCAUUUA ......(((((..((.....))........((((((((((.....))))))..)))).......)))))......---(((.-..((((((...((....)).))))))..)))...... ( -35.60) >DroYak_CAF1 7846 119 + 1 UAUCCUGGCAGCCGCUUAAUGCAAUCAAAAUGCCUCUGGCCGAGUGCCGGAAUGGCAAUUACUACUGCCACAGUUACUGCUG-CACUUCUGCUACCUAUGGCACAGCAUAAAGCCAUUUA .....((((((..((.....))........((((((((((.....))))))..)))).......))))))..(((..(((((-......(((((....))))))))))...)))...... ( -33.40) >consensus UAUCCAGGCAGCCGCUUAAUGCAAUCAAAAUGCCUCUGGCCGAGUGCCGGAAUGGCAAUUACUACAGCCACAGUU___ACUG_CACGUCUGCUACCUAUGGCACAGAAUAAAGCCAUUUA ......(((((..((.....))........((((((((((.....))))))..)))).............(((......)))......)))))....(((((..........)))))... (-22.15 = -22.75 + 0.60)

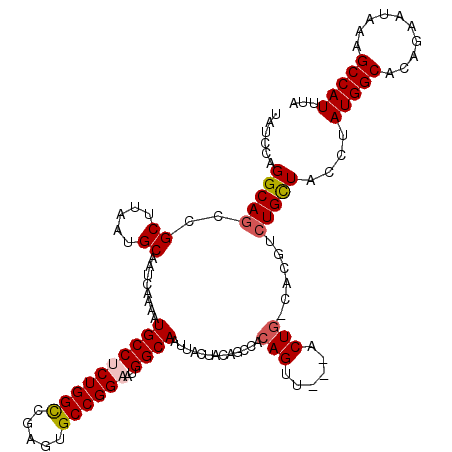
| Location | 18,670,525 – 18,670,641 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.60 |
| Mean single sequence MFE | -40.49 |
| Consensus MFE | -22.71 |
| Energy contribution | -24.77 |
| Covariance contribution | 2.06 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.553701 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18670525 116 - 23771897 UAAAUGGCUUUAUUCUGUGCCAUAGGUAGCAGACGUG-CAGU---AACUGUGGCUGUAGUAAUUGCCAUUCCGGCACUCAGCCAGAGGCAUUUUGAUUGCAUUAAGCGGCUGCCUGGACA ...(((((..........)))))(((((((.(..(((-((((---..((.((((((.(((....(((.....)))))))))))).))........)))))))....).)))))))..... ( -41.60) >DroSec_CAF1 7790 116 - 1 UAAAUGGCUUUAUUCAGUGCCAUAGGUAGCAGACGUG-CAGU---AACUGUGGCUGUAGUAAUUGCCAUUCCGGCACUCGGCCAGCGGCAUUUUGAUUGCAUUAAGCGGCUGCCUGGAUA ...(((((..........)))))(((((((.(..(((-((((---..(((((((((.(((....(((.....)))))))))))).))).......)))))))....).)))))))..... ( -39.80) >DroAna_CAF1 7376 93 - 1 -------------------------GUAGCAGCUGUUCCAGC--UGCCUUUUGCAACGGCAAUUGCCAUUCCGGCACUCGACCAGUGGCAUUUUGAUUGCAUUAAGCGGGUGCCUGGAUA -------------------------((((((((((...))))--)))....)))...(((....)))..(((((((((((.......(((.......)))......)))))))).))).. ( -36.72) >DroSim_CAF1 7734 116 - 1 UAAAUGGCUUUAUUCAGUGCCAUAGGUAGCAGACGUG-CAGU---UACUGUGGCUGUAGUAAUUGCCAUUCCGGCAGUCGGCCAGAGGCAUUUUGAUUGCAUUAAGCGGCUGCCUGGAUA ...(((((..........)))))(((((((.(..(((-((((---((((.((((((.....((((((.....)))))))))))).))......)))))))))....).)))))))..... ( -43.10) >DroEre_CAF1 7698 116 - 1 UAAAUGGCUUUCUUCUGGGCCAUGGCUAACAGAAGUG-CAGC---AACUGUGGCAGUAGUAAUUGCCAUUCCGGCACUCGGCCAGAGGCAUUUUGAUUGCAUUAGGCGGCUGCCUGGAUA ......(((..(((((((((....)))..))))))..-.)))---....((((((((....))))))))(((((((..((.((.((.(((.......))).)).))))..)))).))).. ( -41.40) >DroYak_CAF1 7846 119 - 1 UAAAUGGCUUUAUGCUGUGCCAUAGGUAGCAGAAGUG-CAGCAGUAACUGUGGCAGUAGUAAUUGCCAUUCCGGCACUCGGCCAGAGGCAUUUUGAUUGCAUUAAGCGGCUGCCAGGAUA ...(((((..........))))).((((((.((((((-(....((....((((((((....))))))))....)).(((.....))))))))))...(((.....)))))))))...... ( -40.30) >consensus UAAAUGGCUUUAUUCUGUGCCAUAGGUAGCAGACGUG_CAGC___AACUGUGGCAGUAGUAAUUGCCAUUCCGGCACUCGGCCAGAGGCAUUUUGAUUGCAUUAAGCGGCUGCCUGGAUA ...(((((..........(((((((...((..........)).....)))))))..........)))))(((((((.(((.......(((.......)))......))).)))).))).. (-22.71 = -24.77 + 2.06)

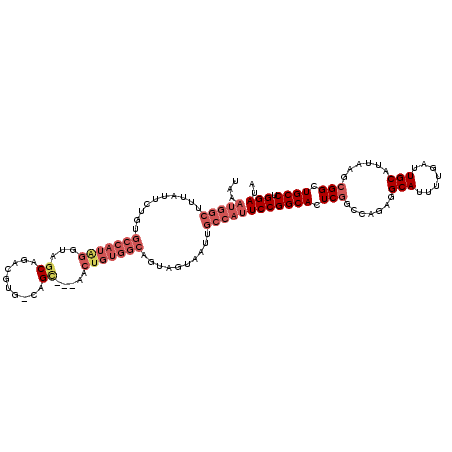
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:33:04 2006