
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 2,080,752 – 2,080,872 |
| Length | 120 |
| Max. P | 0.600185 |

| Location | 2,080,752 – 2,080,872 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.56 |
| Mean single sequence MFE | -38.99 |
| Consensus MFE | -24.44 |
| Energy contribution | -24.87 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.39 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.600185 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2080752 120 - 23771897 CCGGCACCUACACUUGCCGCGCCACCAACAAGCAUGGAACUGCUGAAAUAAGCUGCAAUCUGGAGUGCGUGGACAAGCCGAGGGGCCAGAAGCCGCGCUUCACGUCCCACAUUCAGCCGC .((((.......(((((((((((.(((....(((.......(((......))))))....))).).)))))).))))....(((((..(((((...)))))..))))).......)))). ( -42.61) >DroVir_CAF1 158494 120 - 1 CUGGCACCUACACCUGCCGUGCCACUAACAAAUAUGGUUCGGCUGAGAUAAGCUGCACUUUGGAGUGUGUAGAGAAGCCACGUGGUCAGAAGCCACACUUCACAUCGCAUAUUCAACCGC .((((((...........))))))...........(((((((((......))))).......((((((((.((((((....(((((.....))))).))))...)))))))))))))).. ( -32.60) >DroGri_CAF1 133091 120 - 1 CUGGCACCUACACCUGCCGGGCAACCAAUAAAUAUGGCUCGGCUGAGAUCAGCUGUACGCUGGAGUGUGUGGACAAGCCACGUGGCCAGAAGCCCCACUUCACAUCGCACAUUCAGCCAU ..((((........))))((....)).......((((((((((((....)))))).......((((((((((....(((....)))..((((.....))))...)))))))))))))))) ( -45.60) >DroWil_CAF1 177272 120 - 1 CCGGCACCUAUACCUGCCGCGCAACCAACAAGUACGGAACAGCUGAAAUUAGCUGCACUCUGGAGUGCGUUGAUAAGCCACGUGGACAAAAACCACGUUUCACUUCGCAUCCCCAACCGC .(((((........)))))...............(((..((((((....))))))......((..((((.(((......((((((.......)))))).)))...))))..))...))). ( -32.80) >DroMoj_CAF1 136922 120 - 1 CCGGCACAUACACCUGCCGUGCCACCAACAAAUACGGCUCAGCUGAGAUCAGCUGCACGCUGGAGUGUGUCGACAAGCCUCGUGGCCAGAAACCGCACUUCACGUCGCACAUUCAGCCGC ..(((((...........)))))...........(((((((((((....)))))).......(((((((.((((..(((....)))..(((.......)))..)))))))))))))))). ( -44.50) >DroAna_CAF1 139234 120 - 1 CCGGUACCUACACUUGCCGUGCCACAAAUAAAUACGGCACCGCCGAGACCAGCUGCACUUUGGAGUGCGUGGAUAAGCCGAGGGGCCAGAAACCACGCUUCACCUCUCACAUCCAGCCGC .((((..........((.(((((............))))).)).((((..(((.(((((....)))))((((....(((....)))......))))))).....)))).......)))). ( -35.80) >consensus CCGGCACCUACACCUGCCGUGCCACCAACAAAUACGGCACAGCUGAGAUCAGCUGCACUCUGGAGUGCGUGGACAAGCCACGUGGCCAGAAACCACACUUCACAUCGCACAUUCAGCCGC .(((((........)))))...............((((.((((((....)))))).....(((((((((.......(((....)))..((((.....))))....))))).)))))))). (-24.44 = -24.87 + 0.42)

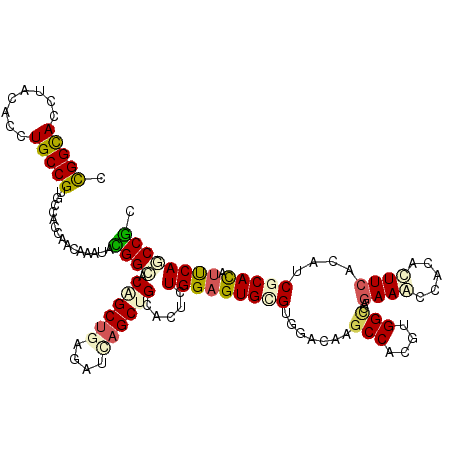
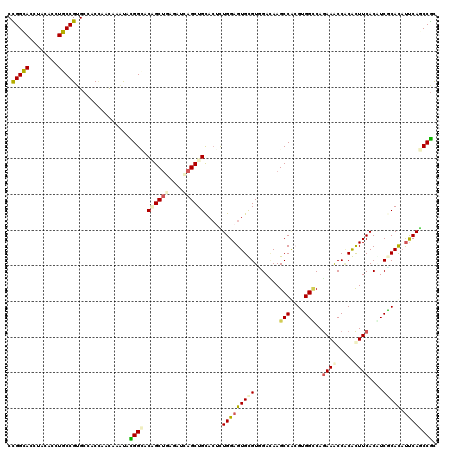
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:48:40 2006