
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 18,669,820 – 18,670,033 |
| Length | 213 |
| Max. P | 0.914220 |

| Location | 18,669,820 – 18,669,929 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 91.53 |
| Mean single sequence MFE | -19.63 |
| Consensus MFE | -18.66 |
| Energy contribution | -18.42 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.914220 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
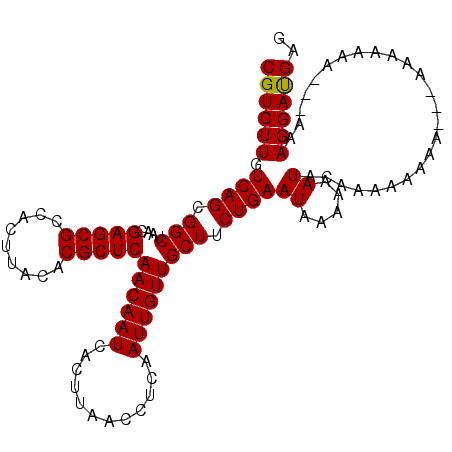
>3L_DroMel_CAF1 18669820 109 + 23771897 CGUCUUGUCAGCGGCUAACGAGCGCCACUUACACGCUCAACAAUCACUUAACCUCAAUUGUUGCUUCUGAAUAAAAAAUACAAAAAAAAAGGAAAAAAA---UAAGGAUGAG ((((((.((((.(((....(((((.........)))))((((((............))))))))).))))..........(.........)........---..)))))).. ( -18.10) >DroSec_CAF1 7098 109 + 1 CGUCUUGUCAGCGGCUAACGAGCGCCACUUACACGCUCAACAAUCACUUAACCUCAAUUGUUGCUUCUGAAUAAAAAAUACAAAAAAAGCAAAAAAAAA---AAAGGAUGAG ((((((.((((.(((....(((((.........)))))((((((............))))))))).))))((.....))....................---..)))))).. ( -17.70) >DroSim_CAF1 7044 104 + 1 CGUCUUGUCAGCGGCUAACGAGCGCCACUUACACGCUCAACAAUCACUUAACCUCAAUUGUUGCUUCUGAAUAAAAAAUACAAAAAA-G----UAAAAA---AAAGGAUGAG ((((((.((((.(((....(((((.........)))))((((((............))))))))).))))........(((......-)----))....---..)))))).. ( -21.40) >DroEre_CAF1 7036 104 + 1 CGUCUUGUCAGCGGCUAACGAGCGCCACUUACACGCUCAACAAUCACUUAACCUCAAUUGUUGCUUCUGAAUAAAAAAUAUAAAAAA-----GGGCGAA---AAAGGACGAG ((((((((.((.(((........))).)).)).(((((((((((............)))))).(....)..................-----)))))..---..)))))).. ( -21.10) >DroYak_CAF1 7146 112 + 1 CGUCUUGUCAGCGGCUAACGAGCGCCACUUACACGCUCAACAAUCACUUAACCUCAAUUGUUGCUUCUGAAUAAAAAAUACAAAAAAAAAAAAGGAAAAAUGAAAGGACGAG ((((((.((((.(((....(((((.........)))))((((((............))))))))).))))..........((..................))..)))))).. ( -19.87) >consensus CGUCUUGUCAGCGGCUAACGAGCGCCACUUACACGCUCAACAAUCACUUAACCUCAAUUGUUGCUUCUGAAUAAAAAAUACAAAAAAAA___AAAAAAA___AAAGGAUGAG ((((((.((((.(((....(((((.........)))))((((((............))))))))).))))((.....)).........................)))))).. (-18.66 = -18.42 + -0.24)


| Location | 18,669,929 – 18,670,033 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.28 |
| Mean single sequence MFE | -33.94 |
| Consensus MFE | -21.95 |
| Energy contribution | -20.94 |
| Covariance contribution | -1.02 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.697619 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18669929 104 - 23771897 CACAAAGGA-----GGAG----------CUGCCACUAAAAU-GAGGGGAGUGCGAAACUAAGAGGUGGAGCCAUUGGCAUUGCUUUUUGCACUUUUCCCUUUACCCUUUGUUGUUGGCCC .(((((((.-----((..----------...)).......(-(((((((((((((......((((..(.(((...))).)..))))))))))...)))))))).)))))))......... ( -32.50) >DroSec_CAF1 7207 104 - 1 CACAAAAGA-----GAAG----------CUGGCAUUAAAAA-GUGGGGGGUGCGAAACUAAGAGGUGGAGCCAUUGGCAUUGCCUUUUGCACUUUUCCCUUUACCCUUUGUUGUUGGCCC .........-----...(----------(..(((.((((..-((((((((........(((((((..(.(((...))).)..))))))).......))))))))..)))).)))..)).. ( -34.26) >DroAna_CAF1 6555 119 - 1 CACAAAAGGCGCAAAAAGGAUCAAGGGAAAGGGAUUCAGAG-AAAGGCAAUGGCAGAGUCAGAGGCAGAGCCAUUGGCAUUGCCUUUUACGCUUUUCCCUUUACCCUUUGUUGUUCUGCU ..........(((..((.(((.((((((((((((....(.(-((((((((((.(((.......(((...))).))).))))))))))).).....))))))).))))).))).)).))). ( -41.60) >DroSim_CAF1 7148 104 - 1 CACAAAAGA-----GAAG----------CUGCCAUUAAAAA-CAGGGGAGUGCGAAACUAAGAGGUGGAGCCAUUGGCAUUGCCUUUUGCACUUUUCCCUUUACCCUUUGUUGUUGGCCC .........-----....----------..((((.....((-(((((((((((((......((((..(.(((...))).)..)))))))))))...........))))))))..)))).. ( -30.10) >DroEre_CAF1 7140 104 - 1 CACAAAAGA-----GAAG----------CUGCCAUUAAAAA-GGGGGCGGGGCGCAACUAAGAGGCGGAGCCAUUGGCAUUGCCUUUUACACUUCUCCCUUUACCCUUUGUUGUUGGCUC .........-----....----------..((((.(((.((-((((..((((.(....((((((((((.(((...))).)))))))))).....).))))...)))))).))).)))).. ( -36.60) >DroYak_CAF1 7258 105 - 1 UACAAAAGA-----GAAG----------CUGCCAUUAAAAAAGAGGGGAGUGCGUAACUAAGAGGUGGAGCCAUUGGCAUUGCCUUUUACACUUUUCCCUUUACCCUUUCUUCUUGGCCC .........-----....----------..((((..((.(((((((((((...((...(((((((..(.(((...))).)..))))))).))..)))))))....)))).))..)))).. ( -28.60) >consensus CACAAAAGA_____GAAG__________CUGCCAUUAAAAA_GAGGGGAGUGCGAAACUAAGAGGUGGAGCCAUUGGCAUUGCCUUUUACACUUUUCCCUUUACCCUUUGUUGUUGGCCC ..............................((((....(((.((((((((........((((((((((.(((...))).)))))))))).....))))))))....))).....)))).. (-21.95 = -20.94 + -1.02)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:33:00 2006