| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 18,667,406 – 18,667,499 |
| Length | 93 |
| Max. P | 0.878614 |

| Location | 18,667,406 – 18,667,499 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.31 |
| Mean single sequence MFE | -20.87 |
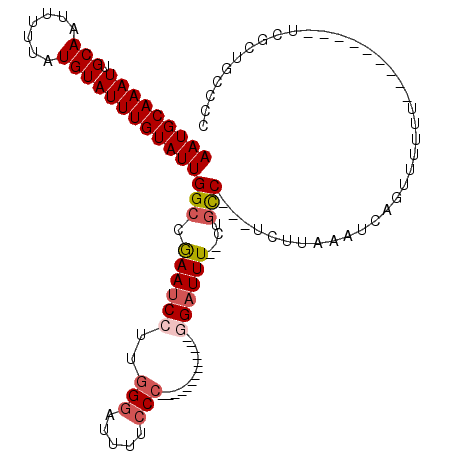
| Consensus MFE | -10.94 |
| Energy contribution | -11.66 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.878614 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
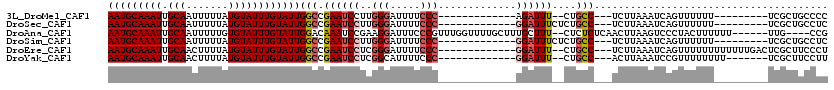
>3L_DroMel_CAF1 18667406 93 - 23771897 AAUGCAAAUUGCAAUUUUUAUGUAUUUGUAUUGGCCGAAUCCUUGGGAUUUUCCC-------------AGAUUU--CUGCC---UCUUAAAUCAGUUUUUU---------UCGCUGCCCC (((((((((.(((.......))))))))))))(((.(((..((.(((.....)))-------------))..))--).)))---........((((.....---------..)))).... ( -21.90) >DroSec_CAF1 4674 95 - 1 AAUGCAAAUUGCAAUUUUUAUGUAUUUGUAUUGGCCGAAUCCUUGGGAUUUUCCC-------------GGAUUUCUCUGCC---UCUUAAAUCAGUUUUUU---------UCGCUGCCUC (((((((((.(((.......))))))))))))(((.((((((..(((.....)))-------------))))))....)))---........((((.....---------..)))).... ( -21.50) >DroAna_CAF1 4358 108 - 1 AAUGCAAAUUGCAAUUUUUGUGUAUUUGUAUUGGACAAAUCCGAAGGAUUUCCCGUUUGGUUUUGCUUUGCUUU--CUCUCUCAACUUAAGUCCCUACUUUUUU------UUG----CCG (((((((((((((.....)))).)))))))))(((((((.(((((((.....)).))))).)))((...))...--..............))))..........------...----... ( -17.80) >DroSim_CAF1 4601 95 - 1 AAUGCAAAUUGCAAUUUUUAUGUAUUUGUAUUGGCCGAAUCCUUGGGAUUUUCCC-------------GGAUUUCUCUGCC---UCUUAAAUCAGUUUUUU---------UCGCUGCCUC (((((((((.(((.......))))))))))))(((.((((((..(((.....)))-------------))))))....)))---........((((.....---------..)))).... ( -21.50) >DroEre_CAF1 4622 102 - 1 AAUGCAAAUUGCAACUUUUAUGUAUUUGUAUUGGCCGAAUCCUCGGGAUUUUCCC-------------GGAUUU--CUGCC---UCUUAAAUCAGUUUUUUUUUUUUGACUCGCUUCCCU (((((((((.(((.......))))))))))))(((.(((..(.((((.....)))-------------))..))--).)))---.................................... ( -22.00) >DroYak_CAF1 4817 95 - 1 AAUGCAAAUUGCAACUUUUAUGUAUUUGUAUUGGCCGAAUCCUCGGCAUUUUCCC-------------GGAUUU--CUGCC---ACUUAAAUCCGUUUUUUUU-------UCGCUUCCUU (((((((((.(((.......)))))))))))).(((((....))))).......(-------------((((((--.....---....)))))))........-------.......... ( -20.50) >consensus AAUGCAAAUUGCAAUUUUUAUGUAUUUGUAUUGGCCGAAUCCUUGGGAUUUUCCC_____________GGAUUU__CUGCC___UCUUAAAUCAGUUUUUU_________UCGCUGCCCC (((((((((.(((.......))))))))))))(((.((((((..(((.....))).............))))))....)))....................................... (-10.94 = -11.66 + 0.72)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:32:58 2006