
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 18,666,625 – 18,666,734 |
| Length | 109 |
| Max. P | 0.744063 |

| Location | 18,666,625 – 18,666,734 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.34 |
| Mean single sequence MFE | -37.52 |
| Consensus MFE | -21.79 |
| Energy contribution | -21.77 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.744063 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18666625 109 - 23771897 AGCCAACUGUUGUUGGCAGCUGGC------GCUGGGACAAUGAACUCU-----CCAGUCGCCACUUGAACCAAGGUAAACCGAUCUAGCUGGCUGAUUUAAUAGCUGCCAAGUCCAAGUA .((((((....))))))....(((------(((((((........)).-----)))).))))(((((......((....))(((...((.(((((......)))))))...)))))))). ( -36.90) >DroSec_CAF1 3887 108 - 1 AGCCAACUGUUGUUGGCAGCUGGC------GCUGGGACAAUGAACUCU-----CCAGUCGCCACUUGAACCAAGGUAAACCGAUCCAGCUGGCUGAUUUAAUUGCUGCC-CGUCCAAGUA .((((((....))))))....(((------(((((((........)).-----)))).))))(((((.((...(((((...((((.((....))))))...)))))...-.)).))))). ( -36.30) >DroAna_CAF1 3508 101 - 1 AGCCAACUGCUGUUGGCAGCUGGCACCCUGGCCGGGACAAUGAACUCGGCCAAACAGCCGGCACUUGAACCCAGGUAAGCCGAUCUGGCUGGGUCACCCAA------------------- .((((((....)))))).((((((....((((((((........))))))))....))))))...(((.(((((.((........)).)))))))).....------------------- ( -47.10) >DroSim_CAF1 3833 96 - 1 AGCCAACUGUUGUUGGCAGCUGGC------GAUGGGACAAUGAACUCU-----CCAGUCGCCACUUGAACCAAGGUAAACCGAUCUAGCUGGAUGAUUUAAUUGCUG------------- .((((((....))))))(((((((------(((.(((..........)-----)).)))))))......(((((((......))))...)))...........))).------------- ( -29.20) >DroEre_CAF1 3845 108 - 1 AGCCAACUGUUGUUGGCAGCUGGC------GCUGGGACAAUGAACUCU-----CCAGCCGCCACUUGCACCAAGGUAACGCGAUCUAGCUGGCUGAUUUAAUUGCUGCC-UGUCCAAGUG .((((((....))))))....(((------(((((((........)).-----)))).))))(((((.((..(((((..(((((.(((....))).....)))))))))-))).))))). ( -37.50) >DroYak_CAF1 3963 108 - 1 AGCCAACUGUUGUUGGCAGCUGGC------GCUGGGACAACGAACUCU-----CCAGCCGCCACUUGAACCAAGGUAAAGCGAUCUAGCUGCCUGAUUUAAUUGCAACC-UGUCGAAGGG .((((((....)))))).((((((------(((((((........)).-----)))).))))).(((((...((((..(((......)))))))..)))))..))..((-(.....))). ( -38.10) >consensus AGCCAACUGUUGUUGGCAGCUGGC______GCUGGGACAAUGAACUCU_____CCAGCCGCCACUUGAACCAAGGUAAACCGAUCUAGCUGGCUGAUUUAAUUGCUGCC__GUCCAAGU_ .((((((....))))))((((((.......(((((......(....)......)))))....((((......))))........)))))).............................. (-21.79 = -21.77 + -0.03)

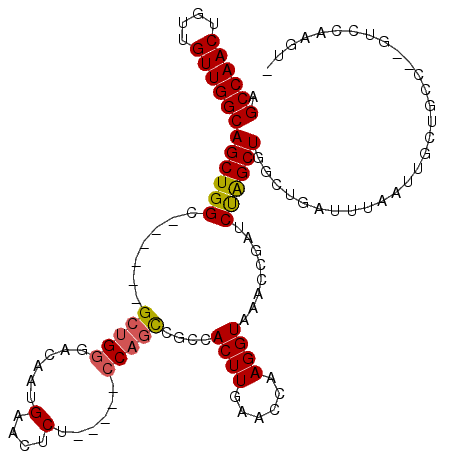

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:32:56 2006