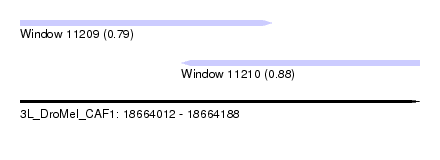
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 18,664,012 – 18,664,188 |
| Length | 176 |
| Max. P | 0.881628 |

| Location | 18,664,012 – 18,664,123 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 92.43 |
| Mean single sequence MFE | -27.88 |
| Consensus MFE | -24.05 |
| Energy contribution | -24.29 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.785556 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18664012 111 + 23771897 UCUGCUGGAGGAGACUAACUUCAAUAUCUGCAGCUCGCUGAUAGCAAGCAUCAACUCCGCACUCAACUUUUACCUUCAAUGCAUUUUACCAGCGACCGCCCAUCGGACAAC ..((((((.((((.....(((...((((.((.....)).))))..)))......))))(((..................)))......)))))).(((.....)))..... ( -23.07) >DroSec_CAF1 1301 111 + 1 UCUGCAGGAGGAGGCUAACUUCAAUAUCUGCAGCUCGCUGAUAGGAAGCAUCAACUCCGCACUGAACUUUUACCUUCAAUGCAUUUUACCAGCGACCGCCCACCGGACAAA .(((((((..((((....))))....))))))).((((((...(((.........)))(((.((((........)))).))).......))))))(((.....)))..... ( -31.00) >DroSim_CAF1 1257 111 + 1 UCUGCAGGAGGAGGCUAACUUCAAUAUCUGCAGCUCGCUGAUAGGAAGCAUCAACUCCGCACUCAACUUUUACCUUCAAUGCAUUUUACCAGCGACCGCCCACCGGACAAC .(((((((..((((....))))....))))))).((((((.(((((.((((...........................)))).))))).))))))(((.....)))..... ( -27.43) >DroEre_CAF1 1246 111 + 1 UCGGCAGGAGGAAGCCAGCUUCAAUAUCUGCUGCUCGCUGAUAGGAAGCAUCGACUCCGCACUCAACUUUUACCUUCAAUGAAUUCUACCAGCGACCGCCCACCGGACACC .(((((((..((((....))))....))))))).((((((.(((((..(((.((.....................)).)))..))))).))))))(((.....)))..... ( -29.40) >DroYak_CAF1 1139 111 + 1 UCGGCAGAAGGAGGCCAACUUCAAUAUCUGCUGCUCGCUGAUAGGAAGCAUCAACUCCGCACUGAACUUUUACCUUCAAUGAAUUUCAGCAGCGACCGCCCACCGGACACC .(((((((..((((....))))....))))))).((((((...(((.........)))...(((((.(((..........))).)))))))))))(((.....)))..... ( -28.50) >consensus UCUGCAGGAGGAGGCUAACUUCAAUAUCUGCAGCUCGCUGAUAGGAAGCAUCAACUCCGCACUCAACUUUUACCUUCAAUGCAUUUUACCAGCGACCGCCCACCGGACAAC .(((((((..((((....))))....))))))).((((((.(((((.((((...........................)))).))))).))))))(((.....)))..... (-24.05 = -24.29 + 0.24)



| Location | 18,664,083 – 18,664,188 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 91.32 |
| Mean single sequence MFE | -36.29 |
| Consensus MFE | -29.42 |
| Energy contribution | -29.86 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.881628 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18664083 105 - 23771897 A-------AACUUAUGCAACUGGGCA-------ACUGAGCUUGUUUAGAUUUAUAAGCUGCCACUGUCCUUGGAUGGGCGUUGUCCGAUGGGCGGUCGCUGGUAAAAUGCAUUGAAGGU .-------..((((((((..((((((-------((..(((((((........)))))))(((...(((....))).)))))))))))((.(((....))).))....)))))..))).. ( -35.10) >DroSec_CAF1 1372 105 - 1 A-------AACUUAUGCAACUGGGCA-------ACUGAGCUUGUUUAGAUUUAUAAGCUGCCACUGUCCGUGGAUGGGCUUUGUCCGGUGGGCGGUCGCUGGUAAAAUGCAUUGAAGGU .-------..((((((((((((((((-------(...(((((((........)))))))(((...(((....))).))).))))))))).(((....))).......)))))..))).. ( -33.70) >DroSim_CAF1 1328 105 - 1 A-------AACUUAUGCAACUGGGCA-------ACUGAGCUUGUUUAGAUUUAUAAGCUGCCACUGUCCUUGGAUGGGCGUUGUCCGGUGGGCGGUCGCUGGUAAAAUGCAUUGAAGGU .-------..((((((((((((((((-------((..(((((((........)))))))(((...(((....))).))))))))))))).(((....))).......)))))..))).. ( -37.70) >DroEre_CAF1 1317 119 - 1 AAACUGCAAACUUAUGCAACUGGGCAACUGGCAACUGAGCUUGUUUAGAUUUAUAAGCUGCCACUGUCCUUGGAUGGGCGGUGUCCGGUGGGCGGUCGCUGGUAGAAUUCAUUGAAGGU ...((((.......(((.(((((((....((((.....((((((........))))))))))(((((((......))))))))))))))..))).......)))).............. ( -39.64) >DroYak_CAF1 1210 105 - 1 A-------AACUUAUGCAGCUGGGCA-------ACUGAGCUUGUUUAGAUUUAUAAGCUGCCACUGUCCUUGGAUGGGCGGUGUCCGGUGGGCGGUCGCUGCUGAAAUUCAUUGAAGGU .-------.......((((...((((-------.....((((((........)))))))))).))))((((.(((((((((((.(((.....))).)))))))......)))).)))). ( -35.30) >consensus A_______AACUUAUGCAACUGGGCA_______ACUGAGCUUGUUUAGAUUUAUAAGCUGCCACUGUCCUUGGAUGGGCGUUGUCCGGUGGGCGGUCGCUGGUAAAAUGCAUUGAAGGU ..........((((((((((((((((...........(((((((........)))))))(((...(((....))).)))..)))))))).(((....))).......)))))..))).. (-29.42 = -29.86 + 0.44)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:32:55 2006