| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 18,661,351 – 18,661,456 |
| Length | 105 |
| Max. P | 0.818747 |

| Location | 18,661,351 – 18,661,456 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 70.15 |
| Mean single sequence MFE | -39.00 |
| Consensus MFE | -12.86 |
| Energy contribution | -13.31 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.33 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.818747 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18661351 105 - 23771897 GAUUGCAUCGACGAUGAGU-UGUGCCUCAGAUGUCUG--------AAGACCUGGGGCAAGGGGUCACAGGUCAGAGGUAGCGGUUUCGAUGUUUUCUGUUCCACUUGCCAGGUU ((..(((((((.(((..((-..((((((((....)).--------..((((((.(((.....))).)))))).)))))))).))))))))))..)).................. ( -33.40) >DroPse_CAF1 1471 104 - 1 GAUUGCAUCCGCAAUGAGGCCUUGCCUCAGCUGGCGGAUAUACGAAAGAUAUGGGGGA-----UCGGCAGCCGUUGGUUGGG-----GCUGUUUUCUGUGUCGGUUGGCAGGUU ..((((((((((..((((((...))))))....))))))...(((..(((((((..((-----.((((..(((.....))).-----))))))..)))))))..)))))))... ( -45.20) >DroSec_CAF1 1244 105 - 1 GAUUGCAUCGACGAUGAGU-UGUGCCUCAGAUGUCUG--------AAGACCUGGGGCAAGGGGUCAGAGGUCAGAGGUAGCGGGUUCGUUGCUUUCUGUCCCACUUGCCAGGUU ....(((.((((.....))-))))).((((....)))--------).(((((..(((((((((.(((((......(((((((....)))))))))))).))).))))))))))) ( -42.20) >DroSim_CAF1 1695 105 - 1 GAUUGCAUCGACGAUGAGU-UGUGCCUCAGAUGUCUG--------AAGACCUGGGGCAAGGGGUCAGAGGUCAGAGGUAGCGGGUUCGUUGCUUUCUGUCCCACUUGCCAGGUU ....(((.((((.....))-))))).((((....)))--------).(((((..(((((((((.(((((......(((((((....)))))))))))).))).))))))))))) ( -42.20) >DroYak_CAF1 1739 81 - 1 GAUUGCAUCGGCGAUGAGU-UGUGCCUCAAAUGUCUG--------CAGGCAUGGGGCAAGGGGUCAGAGGU---------------CGUUGUUUUCU---------GCCAGGUU ((..(((.((((..(((.(-(.(((((((..((((..--------..)))))))))))..)).)))...))---------------)).)))..)).---------........ ( -26.90) >DroPer_CAF1 1487 104 - 1 GAUUGCAUCCGCAAUGAGGCCUUGCCUCAGCUGGCGGAUAUACGAAAGAUAUGGGGGA-----UCGGCAGCCGGUGGUUGGG-----GCUGUUUUCUGUGUCGGUUGGCAGGUU ..((((((((((..((((((...))))))....))))))...(((..(((((((..((-----.((((..(((.....))).-----))))))..)))))))..)))))))... ( -44.10) >consensus GAUUGCAUCGACGAUGAGU_UGUGCCUCAGAUGUCUG________AAGACAUGGGGCAAGGGGUCAGAGGUCAGAGGUAGCG____CGUUGUUUUCUGUCCCACUUGCCAGGUU ((..(((.((((..(((.(...(((((((...((((..........)))).)))))))...).)))...))...............)).)))..)).................. (-12.86 = -13.31 + 0.45)


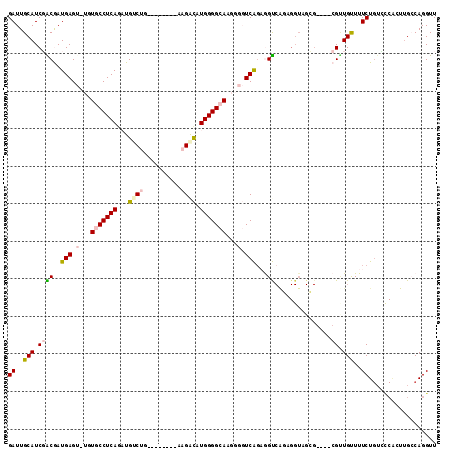
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:32:53 2006