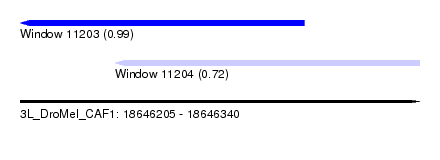
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 18,646,205 – 18,646,340 |
| Length | 135 |
| Max. P | 0.993063 |

| Location | 18,646,205 – 18,646,301 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 70.49 |
| Mean single sequence MFE | -28.46 |
| Consensus MFE | -10.78 |
| Energy contribution | -11.58 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.38 |
| SVM decision value | 2.37 |
| SVM RNA-class probability | 0.993063 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18646205 96 - 23771897 UCAGUCACCAAUGUAUUCUAAACUAAUGGGAUUUUGGUGAUCGACCCCUACUCUUCUUUGAGAU---------------------CAUUUGAAUAGGACAGC---AGUUAGUGGCAAUAU ((.((((((((...((((((......)))))).)))))))).))..(((((((......))).(---------------------(....)).))))...((---........))..... ( -21.70) >DroSec_CAF1 7932 120 - 1 UCAGUCACCAAAGUAUUCUAAACUAAUGGGAUUUUGGUGAUCGACUCCUGCUCUUAUUUUAGAUGAUUACUAGGUGUCUAGUAACCAUUUGAAUAGGAAUGAAGCAGUUAGUGGUAAUAU ...(((((((((..((((((......)))))))))))))))(.(((.((((((((((((.(((((.(((((((....))))))).))))))))))))).....))))..))).)...... ( -35.80) >DroSim_CAF1 7937 117 - 1 UCAGUCACCAAUGUAUUCUAAACUAAUGGGAUUUUGGUGAUCAACUAGAGCUCUUAUUUCAGAUGAUUACUGGUUUUCUAGUAACCAUUUGAAUAGGAAAGA---AGUUAGUGGCAAUAU ...((((((((...((((((......)))))).))))))))..(((((..((((((((.((((((.(((((((....))))))).)))))))))))))..).---..)))))........ ( -34.70) >DroEre_CAF1 7864 102 - 1 UCAGUCACCAAUGCUUACUAAACUAAUGGGACUUUGGUGAUUGACCGCUGUUCUUAC------------------CACUAGUGACCAUUUGCAUAAAAUAGAAGCAGUUACUAGAAGAUU (((((((((((..((((.........))))...))))))))))).............------------------..((((((((....(((...........)))))))))))...... ( -25.70) >DroYak_CAF1 8341 115 - 1 UCAGUCACCAAUGUUUUCUAAACAAAUGGCACUUUGGUGAUUAACCGCUGGUCUUAUUUCGGAUAAAAAU-----UACUAGUAACCUUUCGUAUAAGAUAAAAGCAGUUAAUGGGAAUCU ...((((((((.((...(((......))).)).))))))))((((.(((.(((((((..((((.......-----............)))).)))))))...))).)))).......... ( -24.41) >consensus UCAGUCACCAAUGUAUUCUAAACUAAUGGGAUUUUGGUGAUCGACCCCUGCUCUUAUUUCAGAU_A__A______UACUAGUAACCAUUUGAAUAGGAAAGAAGCAGUUAGUGGCAAUAU ...((((((((...((((((......)))))).))))))))............................................................................... (-10.78 = -11.58 + 0.80)

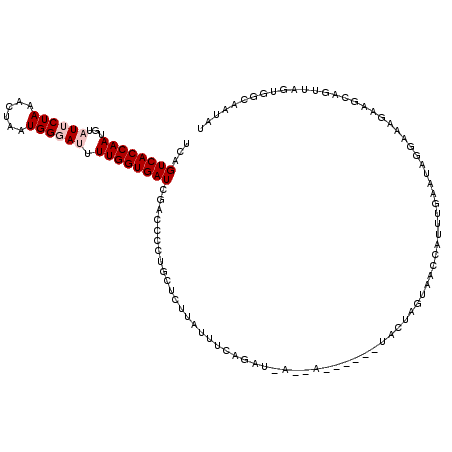

| Location | 18,646,237 – 18,646,340 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 77.25 |
| Mean single sequence MFE | -22.98 |
| Consensus MFE | -13.48 |
| Energy contribution | -14.68 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.718840 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18646237 103 - 23771897 UUUUCGUAGUUUAUUCUGCCAUAAAAGUACACCUUUAAAUCAGUCACCAAUGUAUUCUAAACUAAUGGGAUUUUGGUGAUCGACCCCUACUCUUCUUUGAGAU---------------- .....((((......))))....................((.((((((((...((((((......)))))).)))))))).))......(((......)))..---------------- ( -20.70) >DroSec_CAF1 7972 119 - 1 AUUUCGUAGCUUAUUCUGCCAUAAAAGAACACCUUUAAAUCAGUCACCAAAGUAUUCUAAACUAAUGGGAUUUUGGUGAUCGACUCCUGCUCUUAUUUUAGAUGAUUACUAGGUGUCUA .....((((......))))......(((.(((((...((((((((((((((..((((((......)))))))))))))))..........(((......))))))))...)))))))). ( -28.10) >DroSim_CAF1 7974 115 - 1 ----CGUAGUUUAUUCUGCCAUAAAAGUACACCUUUAAAUCAGUCACCAAUGUAUUCUAAACUAAUGGGAUUUUGGUGAUCAACUAGAGCUCUUAUUUCAGAUGAUUACUGGUUUUCUA ----.((((((...((((..((((.((.....(((((.....((((((((...((((((......)))))).))))))))....))))))).))))..)))).)))))).......... ( -24.60) >DroEre_CAF1 7904 101 - 1 UGUUCGUAGUUUAUUCUGCCAUAAAAGUACACUUUUAAAUCAGUCACCAAUGCUUACUAAACUAAUGGGACUUUGGUGAUUGACCGCUGUUCUUAC------------------CACUA .....((((......))))..((((((....))))))..(((((((((((..((((.........))))...))))))))))).............------------------..... ( -21.70) >DroYak_CAF1 8381 114 - 1 UUUUCGUAGUUUAUUCUGCCAUAAAAGUACACUCUUAAAUCAGUCACCAAUGUUUUCUAAACAAAUGGCACUUUGGUGAUUAACCGCUGGUCUUAUUUCGGAUAAAAAU-----UACUA .....(((((((..((((..((((.................(((((((((.((...(((......))).)).))))))))).(((...))).))))..))))...))))-----))).. ( -19.80) >consensus UUUUCGUAGUUUAUUCUGCCAUAAAAGUACACCUUUAAAUCAGUCACCAAUGUAUUCUAAACUAAUGGGAUUUUGGUGAUCGACCCCUGCUCUUAUUUCAGAU_A__A______UACUA .....((((......))))....................((.((((((((...((((((......)))))).)))))))).)).................................... (-13.48 = -14.68 + 1.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:32:49 2006