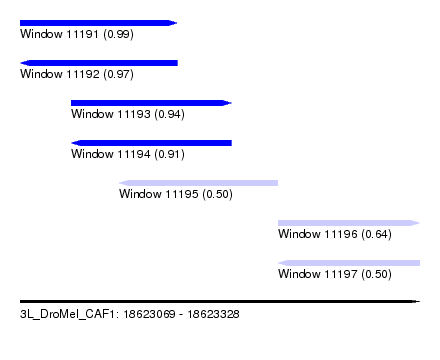
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 18,623,069 – 18,623,328 |
| Length | 259 |
| Max. P | 0.994739 |

| Location | 18,623,069 – 18,623,171 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 75.45 |
| Mean single sequence MFE | -26.64 |
| Consensus MFE | -15.68 |
| Energy contribution | -16.20 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.59 |
| SVM decision value | 2.51 |
| SVM RNA-class probability | 0.994739 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
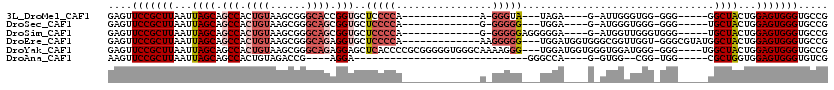
>3L_DroMel_CAF1 18623069 102 + 23771897 CACCACUGUAAACCACGUAC-------GUACGGCCAAAACCCCCAAAUUCCCCAC--AUCCCCUCUUGGC-GGGUGCUAAUGCACUUGCACUCAGUGCAAGCACAUAAUUCG ............((.(((..-------..)))(((((..................--........)))))-))(((((..((((((.......)))))))))))........ ( -21.97) >DroSec_CAF1 34781 106 + 1 CAC---GGCAAACCACCACCCCCUCUUGGGCGGCAA--AC-CCCAAAUCCCCCACACACCCCCUCUUGGCUGGGUGCUAAUGCACUUGCACUCAGUGCAAGCACAUAAUUCG ...---.(((........(((((....))).))...--..-...............(((((((....))..)))))....))).(((((((...)))))))........... ( -26.60) >DroSim_CAF1 35695 107 + 1 CAC---GGCAAACCACCACCCCCUCUUGGGCGGCCA--ACCCCCAAAUCCCCCACACACCCCCUCUUGGCUGGGUGCUAAUGCACUUGCACUCAGUGCAAGCACAUAAUUCG ...---.(((...............(((((.((...--.)))))))..........(((((((....))..)))))....))).(((((((...)))))))........... ( -28.20) >DroEre_CAF1 35405 92 + 1 CAC---AGCCAA-------CCCCUCUGCGGCGCCCC--GC-CCCCCA----CCAC--CACCCCCCUUGGC-GGGUGCUAAUGCACUUGCACUCGGUGCAAGCACAUAAUUCG ...---.(((..-------.........)))(((((--((-(.....----....--..........)))-))).))..(((..(((((((...)))))))..)))...... ( -27.01) >DroYak_CAF1 35753 89 + 1 CAC---CACC-A-------CCCCUUUGGGGAAGGC----C-CCCCAA----CCAC--CAACCCUCUUGGC-GGGUGCUAAUGCACUUGCACUCAGUGCAAGCACAUAAUUCG ...---....-.-------.....((((((.....----.-))))))----((.(--(((.....)))).-))(((((..((((((.......)))))))))))........ ( -29.40) >consensus CAC___GGCAAACCAC___CCCCUCUGGGGCGGCCA__AC_CCCAAAU_CCCCAC__AACCCCUCUUGGC_GGGUGCUAAUGCACUUGCACUCAGUGCAAGCACAUAAUUCG ..........................((((.(.......).))))................((........))(((((..((((((.......)))))))))))........ (-15.68 = -16.20 + 0.52)



| Location | 18,623,069 – 18,623,171 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 75.45 |
| Mean single sequence MFE | -39.38 |
| Consensus MFE | -21.06 |
| Energy contribution | -21.50 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.53 |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.969109 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18623069 102 - 23771897 CGAAUUAUGUGCUUGCACUGAGUGCAAGUGCAUUAGCACCC-GCCAAGAGGGGAU--GUGGGGAAUUUGGGGGUUUUGGCCGUAC-------GUACGUGGUUUACAGUGGUG .(((((((((((((((((...))))))((((.......(((-((...........--))))).........(((....)))))))-------)))))))))))......... ( -30.40) >DroSec_CAF1 34781 106 - 1 CGAAUUAUGUGCUUGCACUGAGUGCAAGUGCAUUAGCACCCAGCCAAGAGGGGGUGUGUGGGGGAUUUGGG-GU--UUGCCGCCCAAGAGGGGGUGGUGGUUUGCC---GUG (((((((((..(((((((...)))))))..)))..((((((..((....))))))))......)))))).(-((--(..((((((......))))))..)...)))---... ( -45.60) >DroSim_CAF1 35695 107 - 1 CGAAUUAUGUGCUUGCACUGAGUGCAAGUGCAUUAGCACCCAGCCAAGAGGGGGUGUGUGGGGGAUUUGGGGGU--UGGCCGCCCAAGAGGGGGUGGUGGUUUGCC---GUG (((((((((..(((((((...)))))))..)))..((((((..((....))))))))......))))))..(((--(.(((((((......))))))).)...)))---... ( -44.60) >DroEre_CAF1 35405 92 - 1 CGAAUUAUGUGCUUGCACCGAGUGCAAGUGCAUUAGCACCC-GCCAAGGGGGGUG--GUGG----UGGGGG-GC--GGGGCGCCGCAGAGGGG-------UUGGCU---GUG ......(((..(((((((...)))))))..)))((((.(((-((((.........--.)))----))))..-((--((....)))).......-------...)))---).. ( -37.00) >DroYak_CAF1 35753 89 - 1 CGAAUUAUGUGCUUGCACUGAGUGCAAGUGCAUUAGCACCC-GCCAAGAGGGUUG--GUGG----UUGGGG-G----GCCUUCCCCAAAGGGG-------U-GGUG---GUG ........(..(((((((...)))))))..)((((.(((((-(((((.....)))--))..----((((((-(----....)))))))..)))-------)-).))---)). ( -39.30) >consensus CGAAUUAUGUGCUUGCACUGAGUGCAAGUGCAUUAGCACCC_GCCAAGAGGGGGU__GUGGGG_AUUUGGG_GU__UGGCCGCCCAAGAGGGG___GUGGUUUGCC___GUG .(....(((..(((((((...)))))))..)))...).(((........))).............((((((.(.......).))))))........................ (-21.06 = -21.50 + 0.44)



| Location | 18,623,102 – 18,623,206 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 88.27 |
| Mean single sequence MFE | -32.30 |
| Consensus MFE | -26.88 |
| Energy contribution | -26.77 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.79 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.939328 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18623102 104 + 23771897 CCCCAAAUUCCCCAC--AUCCCCUCUUGGC-GGGUGCUAAUGCACUUGCACUCAGUGCAAGCACAUAAUUCGCUGCCCUCGAGCACACAUGCAAGAGCAGCCGGCAC ...............--.........((.(-((.((((..(((((((((((...)))))))..........(((.......))).....))))..)))).))).)). ( -30.60) >DroSec_CAF1 34816 106 + 1 -CCCAAAUCCCCCACACACCCCCUCUUGGCUGGGUGCUAAUGCACUUGCACUCAGUGCAAGCACAUAAUUCGCUGCCCUCGAGCACACAUGCGAGAGCAGCCGACAC -...............(((((((....))..)))))...(((..(((((((...)))))))..)))...(((((((.((((..........)))).)))).)))... ( -32.10) >DroSim_CAF1 35730 107 + 1 CCCCAAAUCCCCCACACACCCCCUCUUGGCUGGGUGCUAAUGCACUUGCACUCAGUGCAAGCACAUAAUUCGCUGCCCUCGAGCACACAUGCGAGAGCAGCCGACAC ................(((((((....))..)))))...(((..(((((((...)))))))..)))...(((((((.((((..........)))).)))).)))... ( -32.10) >DroEre_CAF1 35433 99 + 1 -CCCCCA----CCAC--CACCCCCCUUGGC-GGGUGCUAAUGCACUUGCACUCGGUGCAAGCACAUAAUUCGCUGCCCUCGAGCACACAUGCGAGAGCAGCCGACAC -......----((.(--((.......))).-))(((((..((((((.......))))))))))).....(((((((.((((..........)))).)))).)))... ( -32.60) >DroYak_CAF1 35778 99 + 1 -CCCCAA----CCAC--CAACCCUCUUGGC-GGGUGCUAAUGCACUUGCACUCAGUGCAAGCACAUAAUUCGCUGCCCUCGAGCACACAUGCGAGAGCAGCCGACAC -......----((.(--(((.....)))).-))(((((..((((((.......))))))))))).....(((((((.((((..........)))).)))).)))... ( -33.80) >DroAna_CAF1 31202 104 + 1 CUCCA--GCCCAGUCCCACCCACUUAUAAC-GGGUGCUAAUGCACUUGCACUCAGUGCAAGCACAUAAUUCGCUGCCCUCGAGCACACAUGCGAGAGCAGCCGACAC .....--.....(((.(((((.........-)))))...(((..(((((((...)))))))..))).....(((((.((((..........)))).))))).))).. ( -32.60) >consensus _CCCAAAUCCCCCAC__ACCCCCUCUUGGC_GGGUGCUAAUGCACUUGCACUCAGUGCAAGCACAUAAUUCGCUGCCCUCGAGCACACAUGCGAGAGCAGCCGACAC .................................(((((..((((((.......))))))))))).....(((((((.(((..(((....)))))).)))).)))... (-26.88 = -26.77 + -0.11)


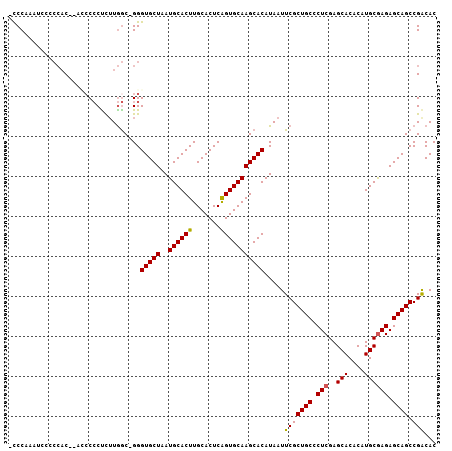
| Location | 18,623,102 – 18,623,206 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 88.27 |
| Mean single sequence MFE | -44.50 |
| Consensus MFE | -36.74 |
| Energy contribution | -36.47 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.905997 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18623102 104 - 23771897 GUGCCGGCUGCUCUUGCAUGUGUGCUCGAGGGCAGCGAAUUAUGUGCUUGCACUGAGUGCAAGUGCAUUAGCACCC-GCCAAGAGGGGAU--GUGGGGAAUUUGGGG ...((.((((((((((((....)))..)))))))))((((((((..(((((((...)))))))..))).....(((-((...........--))))).))))))).. ( -41.00) >DroSec_CAF1 34816 106 - 1 GUGUCGGCUGCUCUCGCAUGUGUGCUCGAGGGCAGCGAAUUAUGUGCUUGCACUGAGUGCAAGUGCAUUAGCACCCAGCCAAGAGGGGGUGUGUGGGGGAUUUGGG- ......((((((((((((....)))..)))))))))((((((((..(((((((...)))))))..)))..((((((..((....))))))))......)))))...- ( -44.90) >DroSim_CAF1 35730 107 - 1 GUGUCGGCUGCUCUCGCAUGUGUGCUCGAGGGCAGCGAAUUAUGUGCUUGCACUGAGUGCAAGUGCAUUAGCACCCAGCCAAGAGGGGGUGUGUGGGGGAUUUGGGG ......((((((((((((....)))..)))))))))((((((((..(((((((...)))))))..)))..((((((..((....))))))))......))))).... ( -44.90) >DroEre_CAF1 35433 99 - 1 GUGUCGGCUGCUCUCGCAUGUGUGCUCGAGGGCAGCGAAUUAUGUGCUUGCACCGAGUGCAAGUGCAUUAGCACCC-GCCAAGGGGGGUG--GUGG----UGGGGG- ......((((((((((((....)))..))))))))).......(..(((((((...)))))))..)((((.(((((-.(....).)))))--.)))----).....- ( -46.50) >DroYak_CAF1 35778 99 - 1 GUGUCGGCUGCUCUCGCAUGUGUGCUCGAGGGCAGCGAAUUAUGUGCUUGCACUGAGUGCAAGUGCAUUAGCACCC-GCCAAGAGGGUUG--GUGG----UUGGGG- ((((..((((((((((((....)))..))))))))).....(((..(((((((...)))))))..)))..))))((-(((((.....)))--))))----......- ( -45.40) >DroAna_CAF1 31202 104 - 1 GUGUCGGCUGCUCUCGCAUGUGUGCUCGAGGGCAGCGAAUUAUGUGCUUGCACUGAGUGCAAGUGCAUUAGCACCC-GUUAUAAGUGGGUGGGACUGGGC--UGGAG ..(((.((((((((((((....)))..))))))))).....(((..(((((((...)))))))..)))...(((((-(.......)))))).))).....--..... ( -44.30) >consensus GUGUCGGCUGCUCUCGCAUGUGUGCUCGAGGGCAGCGAAUUAUGUGCUUGCACUGAGUGCAAGUGCAUUAGCACCC_GCCAAGAGGGGGU__GUGGGGGAUUUGGG_ ((((..((((((((((((....)))..))))))))).....(((..(((((((...)))))))..)))..))))................................. (-36.74 = -36.47 + -0.28)

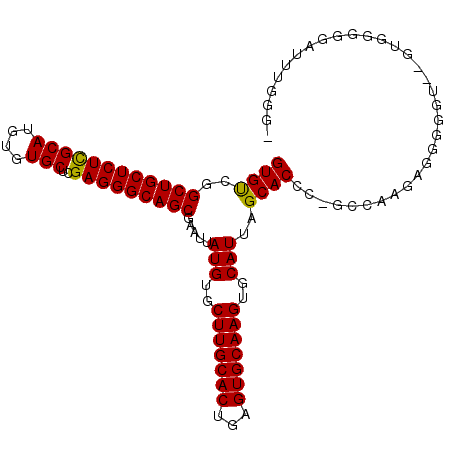

| Location | 18,623,133 – 18,623,236 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.46 |
| Mean single sequence MFE | -45.38 |
| Consensus MFE | -25.67 |
| Energy contribution | -26.08 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.57 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18623133 103 - 23771897 GGUCGCACUGUACUGUAUUGU---------GGCU-CUCGAGUGCCGG-----CUGCUCUUGCAUGUGUGCUCGAGGGCAGCGAAUUAUGUGCUUGCA--CUGAGUGCAAGUGCAUUAGCA ..((((....(((......))---------)(((-((((((..(..(-----((((....))).)))..))))))))).))))...(((..((((((--(...)))))))..)))..... ( -45.70) >DroPse_CAF1 39787 116 - 1 GAACGUCCUCCACU-UGUGGCAUGACGCGGGGCG-UGCGAGUGUCGGUGCCGCCG--CUCUCAUGUGUGCUCGAGGGCAGCGAAUUAUGUGCUUGCCACCGGAGUGCAAGUGCAUUAGCA ....((((((..((-((((......))))))(((-((.(((((.((....)).))--))).)))))......)))))).((.....(((..(((((.((....)))))))..)))..)). ( -47.90) >DroEre_CAF1 35459 102 - 1 GGUCGCACUGUACU-UGUGGC---------GGCU-CUCGAGUGUCGG-----CUGCUCUCGCAUGUGUGCUCGAGGGCAGCGAAUUAUGUGCUUGCA--CCGAGUGCAAGUGCAUUAGCA .((((((.......-))))))---------.(((-((((((..(..(-----((((....))).)))..))))))))).((.....(((..((((((--(...)))))))..)))..)). ( -45.80) >DroYak_CAF1 35804 102 - 1 GGUCCCACUGUACU-UGUGGC---------GGCU-CUCGAGUGUCGG-----CUGCUCUCGCAUGUGUGCUCGAGGGCAGCGAAUUAUGUGCUUGCA--CUGAGUGCAAGUGCAUUAGCA ...(((((......-.)))).---------)(((-((((((..(..(-----((((....))).)))..))))))))).((.....(((..((((((--(...)))))))..)))..)). ( -44.40) >DroAna_CAF1 31233 103 - 1 GGUCGUACUGUACU-UGUGGU---------GGCUGCUCGAGUGUCGG-----CUGCUCUCGCAUGUGUGCUCGAGGGCAGCGAAUUAUGUGCUUGCA--CUGAGUGCAAGUGCAUUAGCA .......(..((((-((.(((---------....)))))))))..)(-----(((((((((((....)))..)))))))))(....(((..((((((--(...)))))))..)))...). ( -40.60) >DroPer_CAF1 40807 116 - 1 GAACGUCCUCCACU-UGUGGCAUGACGCGGGGCG-UGCGAGUGUCGGUGCCGCCG--CUCUCAUGUGUGCUCGAGGGCAGCGAAUUAUGUGCUUGCCACCGGAGUGCAAGUGCAUUAGCA ....((((((..((-((((......))))))(((-((.(((((.((....)).))--))).)))))......)))))).((.....(((..(((((.((....)))))))..)))..)). ( -47.90) >consensus GGUCGCACUGUACU_UGUGGC_________GGCU_CUCGAGUGUCGG_____CUGCUCUCGCAUGUGUGCUCGAGGGCAGCGAAUUAUGUGCUUGCA__CUGAGUGCAAGUGCAUUAGCA ...............................(((.((((((..((((.....)))...........)..))))))))).((.....(((..((((((.......))))))..)))..)). (-25.67 = -26.08 + 0.42)
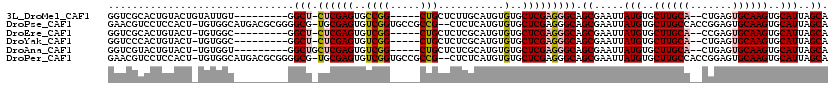
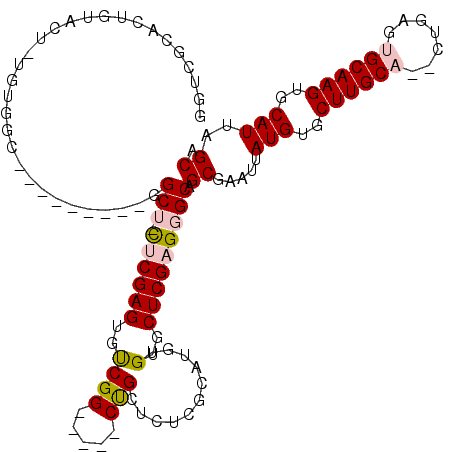
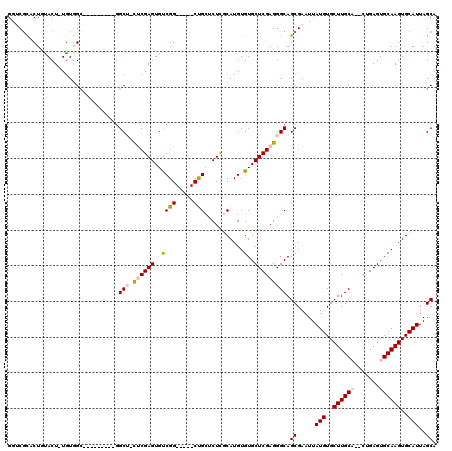
| Location | 18,623,236 – 18,623,328 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 71.98 |
| Mean single sequence MFE | -25.37 |
| Consensus MFE | -13.02 |
| Energy contribution | -13.57 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.641881 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18623236 92 + 23771897 CGGCACCCACUCCAGUAGCC-----CCC-CCACCCAAU-C----UCUA---UACCC-U-------------UGGGGAGCACCGGUGCCCGCUUACAGUGGCUGCUAAUUAAGCGGAACUC .((((((.((....)).((.-----.((-(((......-.----....---.....-.-------------))))).))...))))))((((((.(((....)))...))))))...... ( -26.73) >DroSec_CAF1 34951 92 + 1 CGGCACCCACUCCAGUAGCA-----CCC-CCCACCCAU-C----UCCA---CCCCC-C-------------UGGGGAGCACCGCUGCCCGCUUACAGUGGCUGCUAAUUAAGCGGAACUC ..((((........)).)).-----...-.........-.----(((.---((((.-.-------------.))))((((((((((........)))))).))))........))).... ( -23.80) >DroSim_CAF1 35866 96 + 1 CGGCACCCACUCCAGUAGCA-----CCCACCCAACCAU-C----UCCCCCUCCCCC-C-------------UGGGGAGCACCGCUGCCCGCUUACAGUGGCUGCUAAUUAAGCGGAACUC ......((.((....(((((-----.((((........-.----.....((((((.-.-------------.))))))....((.....)).....)))).)))))....)).))..... ( -26.50) >DroEre_CAF1 35561 103 + 1 CGGCACCCACUCCAGUAGCCAUACGCCC-ACCAACCGCCCACCAUCCA---CCCCCUU-------------UGGGGAGCACCUCUGCCCGCUUACAGUGGCUGCUAAUUAAGCGGAACUC .(((((........)).)))........-.....((((..........---((((...-------------.))))((((((.(((........))).)).))))......))))..... ( -28.30) >DroYak_CAF1 35906 112 + 1 CGGCACCCACUCCAGUAGCCA----CCC-CCCAUCCACCCACCAUCCA---CCCUUUUGCCCACCCCCGCGGGGUGAGCUCCUCUGCCCGCUUACAGUGGCUGCUAAUUAAGCGGAACUC ......((.((..((((((((----(..-...................---..........((((((...)))))).((......)).........))))))))).....)).))..... ( -28.10) >DroAna_CAF1 31336 74 + 1 CGACACCCACUCCACCAGCG-----CCA-CCG--CCAC-C----UGGCCC-----------------------------UCCU----CGGUCUACAGUGGCUGCUAAUUAAGCGGAACUU ......((.((.....((((-----(((-(.(--((..-.----.)))((-----------------------------....----.))......))))).))).....)).))..... ( -18.80) >consensus CGGCACCCACUCCAGUAGCC_____CCC_CCCAACCAC_C____UCCA___CCCCC_U_____________UGGGGAGCACCGCUGCCCGCUUACAGUGGCUGCUAAUUAAGCGGAACUC .((((.(((((.....(((.................................((((................)))).((......))..)))...))))).))))............... (-13.02 = -13.57 + 0.56)
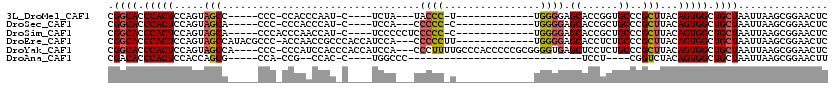
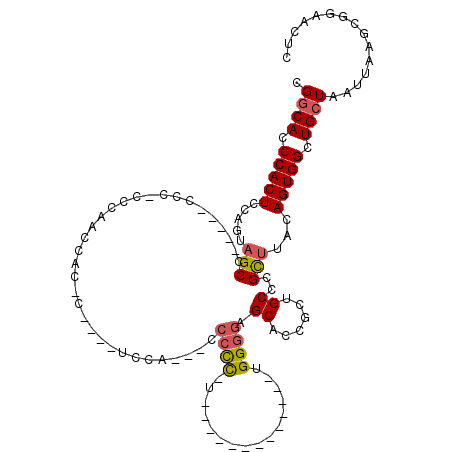
| Location | 18,623,236 – 18,623,328 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 71.98 |
| Mean single sequence MFE | -37.72 |
| Consensus MFE | -17.76 |
| Energy contribution | -19.56 |
| Covariance contribution | 1.79 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.47 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18623236 92 - 23771897 GAGUUCCGCUUAAUUAGCAGCCACUGUAAGCGGGCACCGGUGCUCCCCA-------------A-GGGUA---UAGA----G-AUUGGGUGG-GGG-----GGCUACUGGAGUGGGUGCCG (((.....))).....(((.(((((((......)).((((((((((((.-------------.-....(---(...----.-))......)-)))-----))).)))))))))).))).. ( -33.10) >DroSec_CAF1 34951 92 - 1 GAGUUCCGCUUAAUUAGCAGCCACUGUAAGCGGGCAGCGGUGCUCCCCA-------------G-GGGGG---UGGA----G-AUGGGUGGG-GGG-----UGCUACUGGAGUGGGUGCCG (.(((((((((...(((((.((.(((....))).((.(.((.((((((.-------------.-...))---.)))----)-)).).))..-)).-----)))))...))))))).))). ( -34.40) >DroSim_CAF1 35866 96 - 1 GAGUUCCGCUUAAUUAGCAGCCACUGUAAGCGGGCAGCGGUGCUCCCCA-------------G-GGGGGAGGGGGA----G-AUGGUUGGGUGGG-----UGCUACUGGAGUGGGUGCCG (.(((((((((...(((((.((((((....)...((((.((.((((((.-------------.-.......)))))----)-)).))))))))).-----)))))...))))))).))). ( -41.20) >DroEre_CAF1 35561 103 - 1 GAGUUCCGCUUAAUUAGCAGCCACUGUAAGCGGGCAGAGGUGCUCCCCA-------------AAGGGGG---UGGAUGGUGGGCGGUUGGU-GGGCGUAUGGCUACUGGAGUGGGUGCCG ......((((((.(((((.((((((((..(((((((....)))))(((.-------------...))))---)..))))).))).))))))-)))))...(((.(((......)))))). ( -36.10) >DroYak_CAF1 35906 112 - 1 GAGUUCCGCUUAAUUAGCAGCCACUGUAAGCGGGCAGAGGAGCUCACCCCGCGGGGGUGGGCAAAAGGG---UGGAUGGUGGGUGGAUGGG-GGG----UGGCUACUGGAGUGGGUGCCG ....((((((((.....((.((.((((......))))....(((((((((...)))))))))....)).---)).....))))))))....-.((----((.((((....)))).)))). ( -45.60) >DroAna_CAF1 31336 74 - 1 AAGUUCCGCUUAAUUAGCAGCCACUGUAGACCG----AGGA-----------------------------GGGCCA----G-GUGG--CGG-UGG-----CGCUGGUGGAGUGGGUGUCG ....(((((((.((((((.((((((((..(((.----.((.-----------------------------...)).----)-)).)--)))-)))-----))))))).)))))))..... ( -35.90) >consensus GAGUUCCGCUUAAUUAGCAGCCACUGUAAGCGGGCAGAGGUGCUCCCCA_____________A_GGGGG___UGGA____G_AUGGGUGGG_GGG_____GGCUACUGGAGUGGGUGCCG ....(((((((...((((.(((.((((......)))).)))..(((((................)))))................................))))...)))))))..... (-17.76 = -19.56 + 1.79)


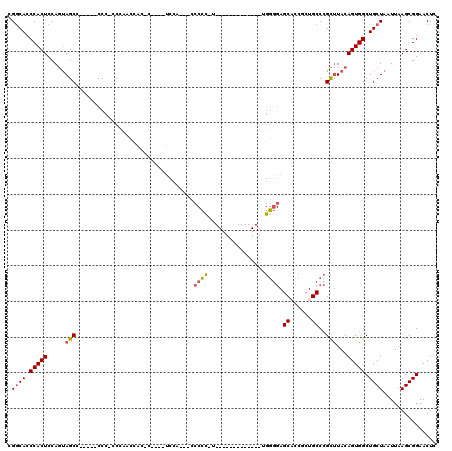
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:32:42 2006