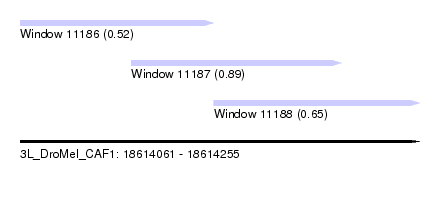
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 18,614,061 – 18,614,255 |
| Length | 194 |
| Max. P | 0.891051 |

| Location | 18,614,061 – 18,614,155 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 79.13 |
| Mean single sequence MFE | -18.79 |
| Consensus MFE | -11.16 |
| Energy contribution | -11.47 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.59 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.517813 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18614061 94 + 23771897 -------UAGAUGCAGACGGAAAUGUCAAACAAAAAUAAGUAGGAAAAGUAGUAGAACCGGAAAUGGGAGAAGACUUGGGGGCAUGGGCAACCAUAUUGGA -------........((((....))))...(((......((.....((((..(....(((....)))...)..))))....))((((....)))).))).. ( -16.10) >DroSec_CAF1 25736 79 + 1 -------UAGAUGCAGACGGAAAUGUCAAACAAAAAUAAGUAGGAAAAGUAGUAG---------------AAGACUUGGUGGCAUGGGUAACCAUAUUGGA -------...((((.((((....))))...((....(((((..............---------------...))))).))))))((....))........ ( -10.43) >DroEre_CAF1 26341 101 + 1 UGAGCGAUAGAUGCAGACGGAAAUGUCAAACAAAAAUAAGUAGGAAAAGCAGUAGAGCCGAAAAUGGGAGAAGGCUUGUGUGCAUGGGCAACCAUAUUGGG ....(((((.((((.((((....)))).....................(((...(((((.............))))).)))))))((....)).))))).. ( -22.02) >DroYak_CAF1 26670 101 + 1 UGAGCGAUAGAUGCAGACGGAAAUGUCAAACAAAAAUGGGUAUGAAAAUCAGUAGAACCAAAAAUGGCAGGAGACUUGUCUGCAUGGGCAACCGUAUUGGA ....(((((.((((((((((...(((((..((....))(((.((.....)).....))).....)))))(....)))))))))))((....)).))))).. ( -26.60) >consensus _______UAGAUGCAGACGGAAAUGUCAAACAAAAAUAAGUAGGAAAAGCAGUAGAACCGAAAAUGGGAGAAGACUUGGGGGCAUGGGCAACCAUAUUGGA ..........((((.((((....))))...................((((.......(((....)))......))))....))))((....))........ (-11.16 = -11.47 + 0.31)

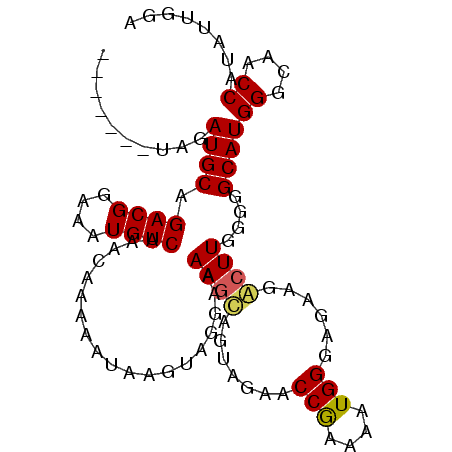
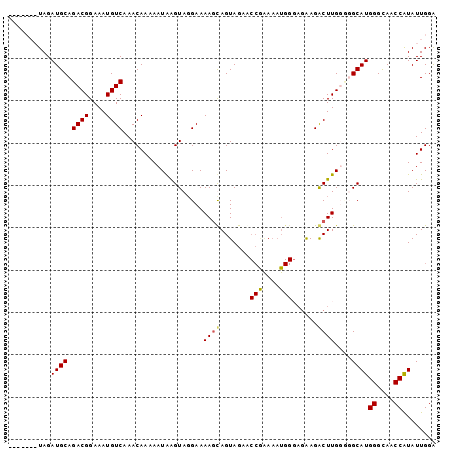
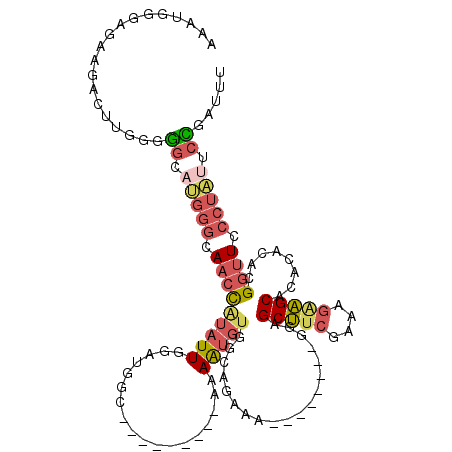
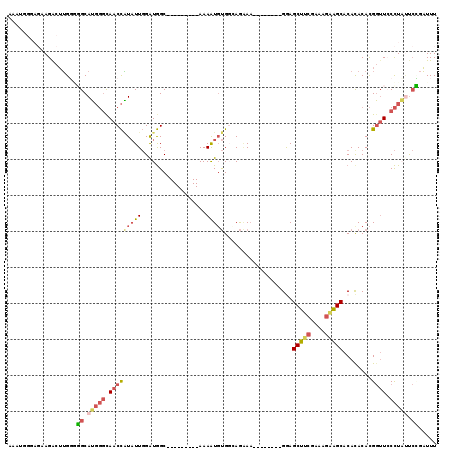
| Location | 18,614,115 – 18,614,217 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 70.67 |
| Mean single sequence MFE | -30.67 |
| Consensus MFE | -13.48 |
| Energy contribution | -14.20 |
| Covariance contribution | 0.73 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.44 |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.891051 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18614115 102 + 23771897 AAAUGGGAGAAGACUUGGGGGCAUGGGCAACCAUAUUGGAUGGC---------AAAAUGUGCCAGAAA--------GGAGCUUCGGCAGAAGCACACACACGGUUCCCUCUUCCGAUUU ...(((((((......((((((((((....))))......((((---------(.....)))))....--------...(((((....))))).........))))))))))))).... ( -35.30) >DroSec_CAF1 25784 93 + 1 ---------AAGACUUGGUGGCAUGGGUAACCAUAUUGGAUGGC---------AAAGUGUGGCAGAAA--------GGAGCUUCGGCAGAAGCACACACACGGUUGCCUAUUCCGAUUU ---------......((((.((....)).)))).((((((.(((---------((.(((((.(.....--------)..(((((....)))))...)))))..)))))...)))))).. ( -30.80) >DroSim_CAF1 25136 102 + 1 AAAUGGGAGAAGACUUGGGGGCAUGGGUAACCAUAUUGGAUGGC---------AAAGUGUGGAAGGAA--------GGAGCUUCGGCAGAAGCACACACACGGUUGCCUAUUCCGAUUU ...............(((..((....))..))).((((((.(((---------((.(((((.......--------.(.(((((....))))).).)))))..)))))...)))))).. ( -31.10) >DroEre_CAF1 26402 101 + 1 AAAUGGGAGAAGGCUUGUGUGCAUGGGCAACCAUAUUGGGUGGC---------AAAAUGUGGGAGAA---------GGAGCUUCGAAGGAAGCACACACACGGUUCCCUAUUCAGAUUU ....(((((..(...(((((((((..((.(((......))).))---------...)))).......---------...(((((....))))))))))..)..)))))........... ( -31.90) >DroYak_CAF1 26731 110 + 1 AAAUGGCAGGAGACUUGUCUGCAUGGGCAACCGUAUUGGAUGGC---------AAAAUGUGGCAGAAAGGCAGAAAGAAGCUUCGAAAGGAGCACAUACACGGUUCCCUACUCAGAUUU .......((....)).(((((..((((.((((((.(((.....)---------))..((((((......))........(((((....))))).)))).)))))).))))..))))).. ( -32.40) >DroAna_CAF1 23283 102 + 1 ---------AAGAAGGGGAGGCAGUUCCAACUUUCUUCGAUAGCGUUUCGCAAAAAAUUCGCAAACAA--------GAAGCCACCAAGAAGGCACACACAUGCAUCCUUCGCAUGUUUU ---------..((((((((((.(((....))).)))))....((((((.((.........))))))..--------...(((........)))........))..)))))......... ( -22.50) >consensus AAAUGGGAGAAGACUUGGGGGCAUGGGCAACCAUAUUGGAUGGC_________AAAAUGUGGCAGAAA________GGAGCUUCGAAAGAAGCACACACACGGUUCCCUAUUCCGAUUU ...................((.(((((.(((((((((..................)))))...................(((((....)))))........)))).))))).))..... (-13.48 = -14.20 + 0.73)
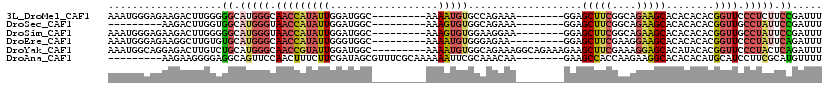
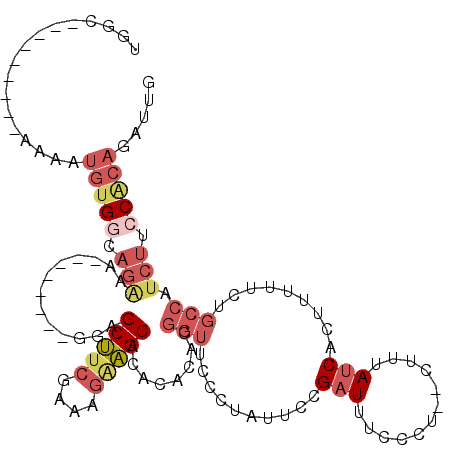
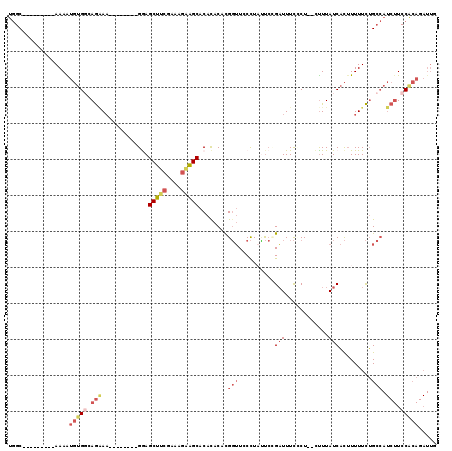
| Location | 18,614,155 – 18,614,255 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 73.86 |
| Mean single sequence MFE | -27.29 |
| Consensus MFE | -11.15 |
| Energy contribution | -12.88 |
| Covariance contribution | 1.73 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.41 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.649595 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18614155 100 + 23771897 UGGC---------AAAAUGUGCCAGAAA--------GGAGCUUCGGCAGAAGCACACACACGGUUCCCUCUUCCGAUUUCGCU--CUUUAUCACUUUUUCUGCCAUCUUCCGCAGAUUG ((((---------(.....)))))((((--------(((((.((((.((((((.(......))))...))).))))....)))--)))).))......(((((........)))))... ( -27.10) >DroSec_CAF1 25815 100 + 1 UGGC---------AAAGUGUGGCAGAAA--------GGAGCUUCGGCAGAAGCACACACACGGUUGCCUAUUCCGAUUUCCCU--CCUUAUCACUUUUUCUGCCAUCUUCCACAGAUUG (((.---------.(((.((((((((((--------((.(((((....))))).......(((.........)))........--.........))))))))))))))))))....... ( -28.00) >DroSim_CAF1 25176 100 + 1 UGGC---------AAAGUGUGGAAGGAA--------GGAGCUUCGGCAGAAGCACACACACGGUUGCCUAUUCCGAUUUCCCU--CUUUAUCACUUUUUCUGCCAUCUUCCACAGAUUG ((((---------((((((...(((((.--------((((((((....))))).......(((.........)))...))).)--))))..)))))....))))).............. ( -28.70) >DroEre_CAF1 26442 99 + 1 UGGC---------AAAAUGUGGGAGAA---------GGAGCUUCGAAGGAAGCACACACACGGUUCCCUAUUCAGAUUUCGCU--CUUUAUCACUUUUUCUGCCAUCUUCCACAGAUUG ...(---------((..(((((((((.---------((.(((((....)))))...................((((.......--.............)))))).)))))))))..))) ( -27.75) >DroYak_CAF1 26771 108 + 1 UGGC---------AAAAUGUGGCAGAAAGGCAGAAAGAAGCUUCGAAAGGAGCACAUACACGGUUCCCUACUCAGAUUUCCCU--CUUUAUCACUUUUUCUGCCAUCUUCCACAGAUUG ...(---------((..(((((.(((..((((((((((.(((((....)))))........((.((........))...))..--.........)))))))))).))).)))))..))) ( -34.40) >DroAna_CAF1 23314 98 + 1 UAGCGUUUCGCAAAAAAUUCGCAAACAA--------GAAGCCACCAAGAAGGCACACACAUGCAUCCUUCGCAUGUUUUUUCUUAUUUUAUCCUGUUUUUGG------UCCA------- ..(((...))).......(((.(((((.--------((.(((........)))....((((((.......))))))..............)).))))).)))------....------- ( -17.80) >consensus UGGC_________AAAAUGUGGCAGAAA________GGAGCUUCGAAAGAAGCACACACACGGUUCCCUAUUCCGAUUUCCCU__CUUUAUCACUUUUUCUGCCAUCUUCCACAGAUUG .................(((((.(((.............(((((....)))))........(((..........(((............))).........))).))).)))))..... (-11.15 = -12.88 + 1.73)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:32:34 2006