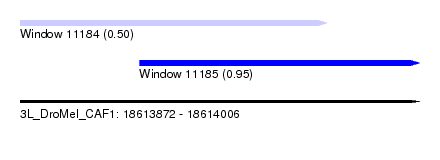
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 18,613,872 – 18,614,006 |
| Length | 134 |
| Max. P | 0.949229 |

| Location | 18,613,872 – 18,613,975 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 87.54 |
| Mean single sequence MFE | -19.48 |
| Consensus MFE | -12.85 |
| Energy contribution | -13.29 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.66 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18613872 103 + 23771897 UGUUUCUCAAAAUAGUUUCUUUCAUUACAAAUGUAUUGUGUGUAAGCAUCGUAAGCACACCUCGCAUUUCGCACAUCAGGUUUUUGCAUAUCGGCAAAUGCCA ............................((((((...((((((..((...))..))))))...)))))).........(((.(((((......))))).))). ( -19.10) >DroSec_CAF1 25547 103 + 1 UUUUUCCCAAAAUAGUUUCUUUCAUUAAAAACUUAUUGUGUGUAAGCAUAGCAAACACACCUCGCAUUUUGCACAUCAGGUUUUUGCAUAUCGGCAAAUGCCA .........................(((((((((...((((((..((...))..))))))...((.....)).....)))))))))......(((....))). ( -19.40) >DroSim_CAF1 24893 103 + 1 UUUUUCCCAAAAUAGUUCCUUUCAUUACAAACUUAUUGUGUGUAAGCAUCGCAAACACACCUCGCAUUUCGCACAUCAGGUUUUUGCAUAUCGGCAAAUGCCA .....................................((((((..((...))..))))))...((.....))......(((.(((((......))))).))). ( -17.90) >DroYak_CAF1 26484 100 + 1 UUUUUGU-AAAAUAUGUUACUUUACUACAAAGGUAUUGUGUGUAUGCAUCAUAAA--CACCUCGCAUUUCGCACAUCAAAUUUUUGCAUAUCGGCAAAUGCCA ....(((-((((.....((((((......)))))).((((((.((((........--......))))..))))))......)))))))....(((....))). ( -21.54) >consensus UUUUUCCCAAAAUAGUUUCUUUCAUUACAAACGUAUUGUGUGUAAGCAUCGCAAACACACCUCGCAUUUCGCACAUCAGGUUUUUGCAUAUCGGCAAAUGCCA ............................((((((..((((((...((................))....))))))..)))))).........(((....))). (-12.85 = -13.29 + 0.44)

| Location | 18,613,912 – 18,614,006 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 92.11 |
| Mean single sequence MFE | -20.82 |
| Consensus MFE | -17.92 |
| Energy contribution | -17.92 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.949229 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18613912 94 + 23771897 UGUAAGCAUCGUAAGCACACCUCGCAUUUCGCACAUCAGGUUUUUGCAUAUCGGCAAAUGCCAUGUGCAGCCCACAUUUCUUUCAAGGUAAAUU (((..((.......((.......)).....((((((..(((.(((((......))))).))))))))).))..))).................. ( -21.10) >DroSec_CAF1 25587 94 + 1 UGUAAGCAUAGCAAACACACCUCGCAUUUUGCACAUCAGGUUUUUGCAUAUCGGCAAAUGCCAUGUGCAGCCCACAUUUCUUUCAAGGUAAAUU (((..((...((...........))....(((((((..(((.(((((......))))).))))))))))))..))).................. ( -21.00) >DroSim_CAF1 24933 94 + 1 UGUAAGCAUCGCAAACACACCUCGCAUUUCGCACAUCAGGUUUUUGCAUAUCGGCAAAUGCCAUGUGCAGCCCACAUUUCUUUCAAGGUAAAUU (((..((...((...........)).....((((((..(((.(((((......))))).))))))))).))..))).................. ( -20.50) >DroEre_CAF1 26194 92 + 1 UGUAUGCGUCGUAAG--CACCUCGCAUUUUGCACACCAGAUUUUUGCAUAUCGGCAAUUGCCAUGUGCAGCCCACAUUUCUUUCAAGGUAAAUU ...(((((..((...--.))..)))))(((((.....(((..(((((((((.(((....))))))))))).....)..)))......))))).. ( -20.50) >DroYak_CAF1 26523 92 + 1 UGUAUGCAUCAUAAA--CACCUCGCAUUUCGCACAUCAAAUUUUUGCAUAUCGGCAAAUGCCAUGUGCAGCCCACAUUCCUUUCAAGGUGAAUU ...............--(((((.((.....))...........((((((((.(((....)))))))))))...............))))).... ( -21.00) >consensus UGUAAGCAUCGUAAACACACCUCGCAUUUCGCACAUCAGGUUUUUGCAUAUCGGCAAAUGCCAUGUGCAGCCCACAUUUCUUUCAAGGUAAAUU ..................((((.((.....))...........((((((((.(((....)))))))))))...............))))..... (-17.92 = -17.92 + -0.00)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:32:31 2006