
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 18,609,528 – 18,609,643 |
| Length | 115 |
| Max. P | 0.666461 |

| Location | 18,609,528 – 18,609,643 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 87.77 |
| Mean single sequence MFE | -26.76 |
| Consensus MFE | -22.40 |
| Energy contribution | -22.32 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.636888 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
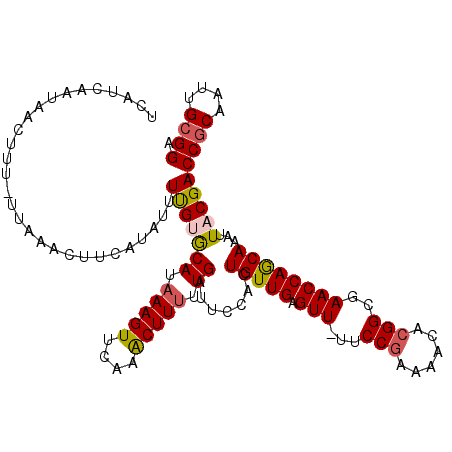
>3L_DroMel_CAF1 18609528 115 + 23771897 UCCGCAAUUGGGGUCGUAAUUUGUUGGUUCGCCGUGUUUUCGGAA-AACUCAACAGUGGAAUCAAAAAGUUUGAACUUUAUGCACAAAAUAUGAAGUUCAAUAAAGUUAUUGAUGA ((((((((((......)))))((((((((..(((......)))..-))).))))))))))(((((.((.(((((((((((((.......)))))))))))...)).)).))))).. ( -30.30) >DroSec_CAF1 21149 114 + 1 UCCGCAAUUGCGGUCGUAAUUUGCUGGUUCGCCGUGUUUUCGGAA-AACUCAACAGUGGAAUCAAAAAGUGUGAACUUUAUGCACGAAAUAUGAAGUUUGA-AAAAUUAUUGAUGA (((((((((((....))))))((.(((((..(((......)))..-))).)).)))))))(((((.((...(((((((((((.......))))))))))).-....)).))))).. ( -27.00) >DroSim_CAF1 20533 113 + 1 UCCGCAAUUGCGGUCGUAAUUUGCUGGUUCGCCGUGUUUUCGGAA-AACUCAACAGUGGAAUCAAAAAGUGUGAACUUUAUGCACAAAAUAUGAAGUUUGA-AAA-GUAUUGUUGA ((((.((..((((.((.((((....))))))))))..)).)))).-...(((((((((....((.....)).((((((((((.......))))))))))..-...-.))))))))) ( -25.20) >DroEre_CAF1 21908 111 + 1 UCCGCAAUUGCGGUCGUAAUUUGCUGGUUCGCCCUGCUUUCGGAA-AACUCAACAGUGGAAUCAAAAAGCUUCAACUUUCUGUUCAAAACAAAAAGUUUAA-AAAGUU---UACAA (((((((((((....))))))((.(((((..((........))..-))).)).)))))))......((((((.((((((.(((.....))).))))))...-.)))))---).... ( -21.50) >DroYak_CAF1 22193 111 + 1 UCCGCAAUUGCGGUCGUAAUUUGCUGGUUUGCCGUGUUUUCGGAAAAACUCAACAGUGGAAUCAA-AAGCUUAAACUUUAUGCUCAAAACAUGAAGUUUAA-AAAGUU---UAUGA (((((((((((....))))))((.((((((.(((......)))..)))).)).))))))).(((.-((((((((((((((((.......))))))))))).-..))))---).))) ( -29.80) >consensus UCCGCAAUUGCGGUCGUAAUUUGCUGGUUCGCCGUGUUUUCGGAA_AACUCAACAGUGGAAUCAAAAAGUUUGAACUUUAUGCACAAAAUAUGAAGUUUAA_AAAGUUAUUGAUGA (((((((((((....))))))((.(((((..(((......)))...))).)).)))))))...........(((((((((((.......)))))))))))................ (-22.40 = -22.32 + -0.08)


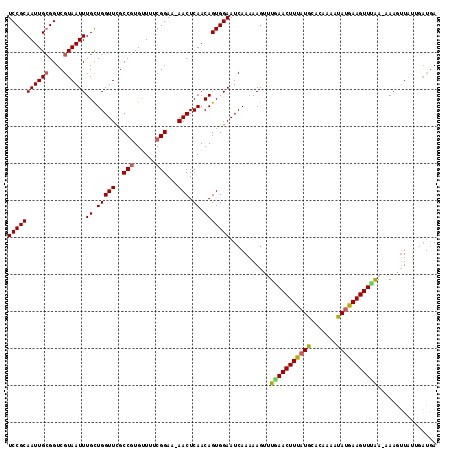
| Location | 18,609,528 – 18,609,643 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 87.77 |
| Mean single sequence MFE | -24.50 |
| Consensus MFE | -17.52 |
| Energy contribution | -17.80 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.666461 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18609528 115 - 23771897 UCAUCAAUAACUUUAUUGAACUUCAUAUUUUGUGCAUAAAGUUCAAACUUUUUGAUUCCACUGUUGAGUU-UUCCGAAAACACGGCGAACCAACAAAUUACGACCCCAAUUGCGGA ..(((((........(((((((((((.....)))....)))))))).....)))))(((..(((((.(((-(.(((......))).))))))))).....(((......))).))) ( -20.22) >DroSec_CAF1 21149 114 - 1 UCAUCAAUAAUUUU-UCAAACUUCAUAUUUCGUGCAUAAAGUUCACACUUUUUGAUUCCACUGUUGAGUU-UUCCGAAAACACGGCGAACCAGCAAAUUACGACCGCAAUUGCGGA ..............-..............(((((((.(((((....))))).)).......(((((.(((-(.(((......))).)))))))))...)))))((((....)))). ( -25.10) >DroSim_CAF1 20533 113 - 1 UCAACAAUAC-UUU-UCAAACUUCAUAUUUUGUGCAUAAAGUUCACACUUUUUGAUUCCACUGUUGAGUU-UUCCGAAAACACGGCGAACCAGCAAAUUACGACCGCAAUUGCGGA ..........-...-..............(((((((.(((((....))))).)).......(((((.(((-(.(((......))).)))))))))...)))))((((....)))). ( -23.00) >DroEre_CAF1 21908 111 - 1 UUGUA---AACUUU-UUAAACUUUUUGUUUUGAACAGAAAGUUGAAGCUUUUUGAUUCCACUGUUGAGUU-UUCCGAAAGCAGGGCGAACCAGCAAAUUACGACCGCAAUUGCGGA (((((---(.((((-...((((((((((.....))))))))))))))..............(((((.(((-(.((........)).)))))))))..))))))((((....)))). ( -28.70) >DroYak_CAF1 22193 111 - 1 UCAUA---AACUUU-UUAAACUUCAUGUUUUGAGCAUAAAGUUUAAGCUU-UUGAUUCCACUGUUGAGUUUUUCCGAAAACACGGCAAACCAGCAAAUUACGACCGCAAUUGCGGA .....---.....(-((((((((.((((.....)))).)))))))))...-..........(((((.((((..(((......))).)))))))))........((((....)))). ( -25.50) >consensus UCAUCAAUAACUUU_UUAAACUUCAUAUUUUGUGCAUAAAGUUCAAACUUUUUGAUUCCACUGUUGAGUU_UUCCGAAAACACGGCGAACCAGCAAAUUACGACCGCAAUUGCGGA .............................(((((((.(((((....))))).)).......(((((.(((...(((......)))..))))))))...)))))((((....)))). (-17.52 = -17.80 + 0.28)

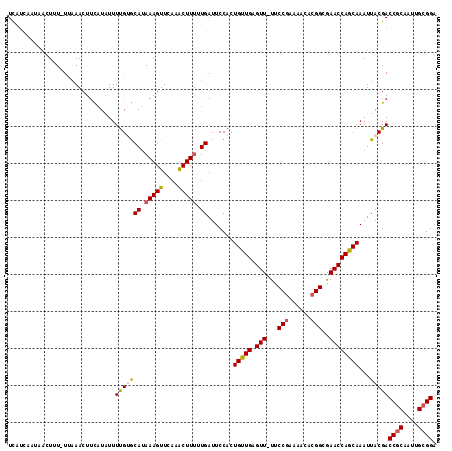
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:32:28 2006