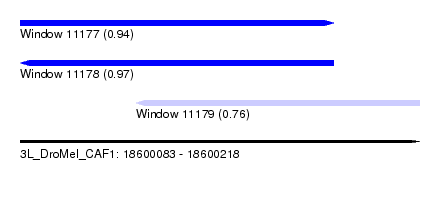
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 18,600,083 – 18,600,218 |
| Length | 135 |
| Max. P | 0.972880 |

| Location | 18,600,083 – 18,600,189 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.12 |
| Mean single sequence MFE | -29.42 |
| Consensus MFE | -19.26 |
| Energy contribution | -19.35 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.95 |
| Structure conservation index | 0.65 |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.935079 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
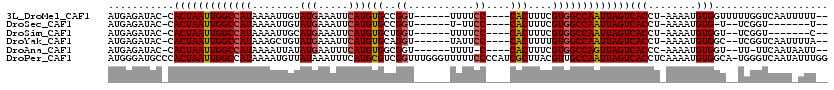
>3L_DroMel_CAF1 18600083 106 + 23771897 AUGAGAUAC-CACUAAUUGGCCAUAAAAUUGUAUGAAAUUCAUGUGCCGGU------UUUUCC----CACUUUCGUGGCCAAUUAGUCACCU-AAAAUGUGGUUUUUGGUCAAUUUUU-- ..(((((((-(((((((((((((..(((.((((((.....))))....((.------....))----)).)))..)))))))))))).(((.-.......)))....)))..))))).-- ( -27.20) >DroSec_CAF1 11748 95 + 1 AUGAGAUAC-CACUAAUUGGCCAUAAAAUUGUAUGAAAUUCAUGUGCCGGU------U-UUCC----CACUUUCGUGGCCAAUUAGUCACCU-AAAAUGUG-U--UCGGU-------U-- .......((-(((((((((((((..(((.((((((.....))))....((.------.-..))----)).)))..))))))))))))(((..-.....)))-.--..)))-------.-- ( -25.10) >DroSim_CAF1 11555 97 + 1 AUGAGAUAC-CACUAAUUGGCCAUAAAAUUGCAUGAAAUUCAUGUGCUGGU------UUUUCC----CACUUUCGUGGCCAAUUAGUCACCU-AAAAUGUGGU--UCGGU-------C-- .......((-(((((((((((((..(((..(((((.....)))))..(((.------.....)----)).)))..)))))))))))).(((.-.......)))--..)))-------.-- ( -30.40) >DroYak_CAF1 12649 104 + 1 AUGAGAUAC-CACUAAUUGGCCAUAAAGCUGUAUGAAAUUCAUGUGCAGGU------UAUUCC----CACUUUUGUGGCCAAUUAGUCACCU-AAAAUGUGGC--UCGGUCAAUUUUA-- .((((((((-(..((((((((((((((((...(((.....)))..)).((.------.....)----)...))))))))))))))(((((..-.....)))))--..)))..))))))-- ( -31.20) >DroAna_CAF1 10530 102 + 1 AUGAGAUAC-CACUAAUUGGCCAUAAAAUUAUAUGAAUUUCAUGUGGCGGU------UUUU-C----CACUUUCGUGGCCAGUUAGUCACCC-AAAAUGUGGU--UU-UUCAAUAAUU-- .(((((.((-(((((((((((((..((((((((((.....))))))).((.------....-)----)..)))..)))))))))))).((..-.....)))))--.)-))))......-- ( -30.90) >DroPer_CAF1 13862 119 + 1 AUGGGAUGCCCACUAAUUGGCCAUAAAAUGUUAUAAAUUUCAUGCGUCGGUUUGGGUUUUUCCCCAUCGCUUACGUUGCCAAUUAGUCACCUCAAAAUGUGGCA-UGGGUCAAUAUUUGG .......(((((.(((((((((((.((((.......)))).)))(((.(((.((((......))))..))).)))..))))))))(((((........))))).-))))).......... ( -31.70) >consensus AUGAGAUAC_CACUAAUUGGCCAUAAAAUUGUAUGAAAUUCAUGUGCCGGU______UUUUCC____CACUUUCGUGGCCAAUUAGUCACCU_AAAAUGUGGU__UCGGUCAAU_UUU__ ...........(((((((((((((........(((.....)))(((..((...........))....)))....)))))))))))))(((........)))................... (-19.26 = -19.35 + 0.09)
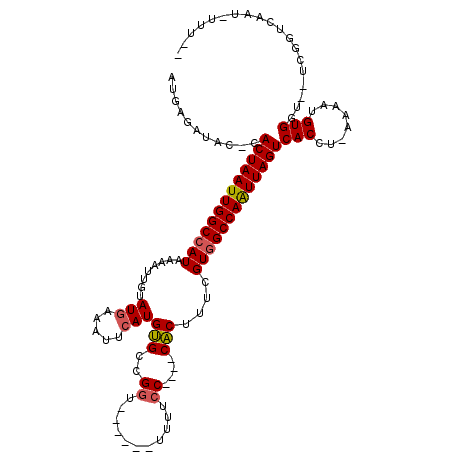
| Location | 18,600,083 – 18,600,189 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.12 |
| Mean single sequence MFE | -31.40 |
| Consensus MFE | -20.10 |
| Energy contribution | -20.38 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.11 |
| Mean z-score | -4.00 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.70 |
| SVM RNA-class probability | 0.972880 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18600083 106 - 23771897 --AAAAAUUGACCAAAAACCACAUUUU-AGGUGACUAAUUGGCCACGAAAGUG----GGAAAA------ACCGGCACAUGAAUUUCAUACAAUUUUAUGGCCAAUUAGUG-GUAUCUCAU --........(((....(((.......-.))).((((((((((((..(((((.----((....------.)).))..(((.....)))....)))..)))))))))))))-))....... ( -30.90) >DroSec_CAF1 11748 95 - 1 --A-------ACCGA--A-CACAUUUU-AGGUGACUAAUUGGCCACGAAAGUG----GGAA-A------ACCGGCACAUGAAUUUCAUACAAUUUUAUGGCCAAUUAGUG-GUAUCUCAU --.-------(((..--.-(((.....-..)))((((((((((((..(((((.----((..-.------.)).))..(((.....)))....)))..)))))))))))))-))....... ( -30.30) >DroSim_CAF1 11555 97 - 1 --G-------ACCGA--ACCACAUUUU-AGGUGACUAAUUGGCCACGAAAGUG----GGAAAA------ACCAGCACAUGAAUUUCAUGCAAUUUUAUGGCCAAUUAGUG-GUAUCUCAU --.-------(((..--(((.......-.))).((((((((((((..(((.((----(.....------.)))...((((.....))))...)))..)))))))))))))-))....... ( -32.10) >DroYak_CAF1 12649 104 - 1 --UAAAAUUGACCGA--GCCACAUUUU-AGGUGACUAAUUGGCCACAAAAGUG----GGAAUA------ACCUGCACAUGAAUUUCAUACAGCUUUAUGGCCAAUUAGUG-GUAUCUCAU --........(((..--(((.......-.))).((((((((((((.(((.(..----((....------.))..)..(((.....))).....))).)))))))))))))-))....... ( -29.90) >DroAna_CAF1 10530 102 - 1 --AAUUAUUGAA-AA--ACCACAUUUU-GGGUGACUAACUGGCCACGAAAGUG----G-AAAA------ACCGCCACAUGAAAUUCAUAUAAUUUUAUGGCCAAUUAGUG-GUAUCUCAU --......(((.-(.--(((((...((-((....)))).((((((..((((((----(-....------....))))(((.....)))....)))..))))))....)))-)).).))). ( -30.60) >DroPer_CAF1 13862 119 - 1 CCAAAUAUUGACCCA-UGCCACAUUUUGAGGUGACUAAUUGGCAACGUAAGCGAUGGGGAAAAACCCAAACCGACGCAUGAAAUUUAUAACAUUUUAUGGCCAAUUAGUGGGCAUCCCAU ........((.(((.-.(((.(.....).))).((((((((((..(((......((((......)))).....)))((((((((.......)))))))))))))))))))))))...... ( -34.60) >consensus __AAA_AUUGACCGA__ACCACAUUUU_AGGUGACUAAUUGGCCACGAAAGUG____GGAAAA______ACCGGCACAUGAAUUUCAUACAAUUUUAUGGCCAAUUAGUG_GUAUCUCAU ...................(((........)))((((((((((((.(((((((....((...........))..)))(((.....)))....)))).))))))))))))........... (-20.10 = -20.38 + 0.28)


| Location | 18,600,122 – 18,600,218 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 79.90 |
| Mean single sequence MFE | -23.87 |
| Consensus MFE | -15.62 |
| Energy contribution | -15.82 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.761938 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18600122 96 - 23771897 UUUUUCUCUGC---------UGGAUUAUAGGGUAUCACAAAAAUUGACCAAAAACCACAUUUUAGGUGACUAAUUGGCCACGAAAGUGGGAAAAACCGGCACAUG ........(((---------(((.......(((.............))).....(((....((((....)))).)))((((....))))......)))))).... ( -25.12) >DroSec_CAF1 11787 85 - 1 UUUUUCUCUGC---------UGGAUUACAGGGUAUCACA-------ACCGA--A-CACAUUUUAGGUGACUAAUUGGCCACGAAAGUGGGAA-AACCGGCACAUG ........(((---------(((.......(((......-------)))..--.-..((..((((....)))).)).((((....))))...-..)))))).... ( -25.30) >DroSim_CAF1 11594 87 - 1 UUUUCCUCUGC---------UGGAUUACAGGGUAUCACG-------ACCGA--ACCACAUUUUAGGUGACUAAUUGGCCACGAAAGUGGGAAAAACCAGCACAUG ........(((---------(((.......(((......-------)))..--.(((....((((....)))).)))((((....))))......)))))).... ( -27.20) >DroEre_CAF1 12517 94 - 1 CUUUCCUCUGC---------UGGAUUAUAGGGUAUCACAAAAAUUGACCAA--ACCUCAUGUUAGGUGACUAAUUGGCCACGAAAGUGGGAAAAACCGGCACAUG ........(((---------(((.......(((.............)))..--.((....(((((....))))).))((((....))))......)))))).... ( -25.82) >DroYak_CAF1 12688 94 - 1 CUUUCCUCUGC---------CGGAUUAUAGGGUAUCACUAAAAUUGACCGA--GCCACAUUUUAGGUGACUAAUUGGCCACAAAAGUGGGAAUAACCUGCACAUG ...(((.....---------.)))...((((.(((..(((...(((...(.--((((....((((....)))).))))).)))...)))..))).))))...... ( -19.80) >DroAna_CAF1 10569 99 - 1 --AUCCCCCGCACACUUCAAUCGACUGAAAGGUAUGAAAAUUAUUGAA-AA--ACCACAUUUUGGGUGACUAACUGGCCACGAAAGUGG-AAAAACCGCCACAUG --.......((....((((......)))).(((...............-..--.(((....((((....)))).)))((((....))))-....)))))...... ( -20.00) >consensus UUUUCCUCUGC_________UGGAUUAUAGGGUAUCACAAAAAUUGACCAA__ACCACAUUUUAGGUGACUAAUUGGCCACGAAAGUGGGAAAAACCGGCACAUG ..............................(((.....................(((....((((....)))).)))((((....)))).....)))........ (-15.62 = -15.82 + 0.19)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:32:25 2006