| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 18,599,649 – 18,599,739 |
| Length | 90 |
| Max. P | 0.754814 |

| Location | 18,599,649 – 18,599,739 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.99 |
| Mean single sequence MFE | -22.50 |
| Consensus MFE | -17.09 |
| Energy contribution | -16.65 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.754814 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
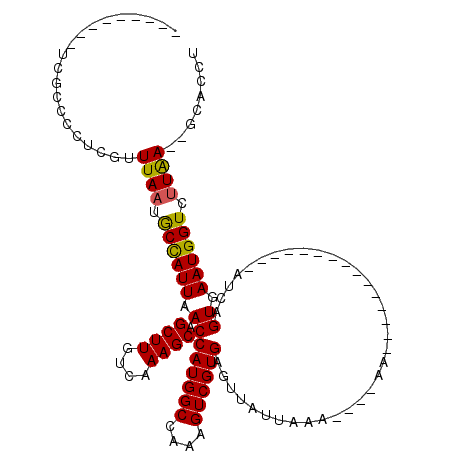
>3L_DroMel_CAF1 18599649 90 - 23771897 ---------UCGCCCCUCGUUUAUUACCAUUAACAGCUUGUCAAAGCCAUGGCCAAAGUCGUGAGUUAUUAAA----AA---------------AUCAGUGAAUGGUCUUAA--GCACCU ---------.........(((((..((((((.((.((((....))))((((((....))))))..........----..---------------....)).))))))..)))--)).... ( -22.20) >DroPse_CAF1 12852 118 - 1 CCCGGCUCUUUGGCACUCGUUUAAGGCUAUUAACAGCUUGUCAAAGCCAUGGCCAAAGUCGUGAGUUAUUAAAGAAAAAGGAGAACAAAAA--AAUCAGUGAAUGGUCUUGAGAGCACCU ....((((((.(((.(((.(((.(((((......))))).....(((((((((....)))))).))).........))).)))........--...((.....)))))..)))))).... ( -26.90) >DroEre_CAF1 12037 90 - 1 ---------CCACCCCUCUUUUAAUGCCAUUAACAGCUUGUCAAAGCCAUGGCCAAAGUCGUGAGUUAUUAAA----AA---------------AUCAGUGAAUGGUCUUAA--GCACCU ---------..........(((((.((((((.((.((((....))))((((((....))))))..........----..---------------....)).)))))).))))--)..... ( -18.80) >DroYak_CAF1 12217 89 - 1 ---------UC-CCCCUCUUUUAAUGCCAUUAACAGCUUGUCAAAGCCAUGGCCAAAGUCGUGAGUUAUUAAA----AA---------------AUCAGUGAAUGGUCUUAA--GCACCU ---------..-.......(((((.((((((.((.((((....))))((((((....))))))..........----..---------------....)).)))))).))))--)..... ( -18.80) >DroAna_CAF1 10118 102 - 1 ------------UCUCUCGUUUAAUGCUAUUAACAGCUUGUCAAAGCCAUGGCCAAAGUCGUGAGUUAUUAAA----AAGGAGAAAAAAAAUACAUCAGUGAAUGGUCUUAA--GCACCU ------------(((((..(((((((((.......((((....))))((((((....)))))))).)))))))----..)))))....................((((....--).))). ( -21.40) >DroPer_CAF1 13377 117 - 1 CCCGGCUCUUUGGCACUCGUUUAAGGCUAUUAACAGCUUGUCAAAGCCAUGGCCAAAGUCGUGAGUUAUUAAAGAAAAAGGAGAACAAA-A--AAUCAGUGAAUGGUCUUGAGAGCACCU ....((((((.(((.(((.(((.(((((......))))).....(((((((((....)))))).))).........))).)))......-.--...((.....)))))..)))))).... ( -26.90) >consensus _________UCGCCCCUCGUUUAAUGCCAUUAACAGCUUGUCAAAGCCAUGGCCAAAGUCGUGAGUUAUUAAA____AA_______________AUCAGUGAAUGGUCUUAA__GCACCU ....................((((.((((((.((.((((....))))((((((....))))))...................................)).)))))).))))........ (-17.09 = -16.65 + -0.44)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:32:22 2006