| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 18,583,621 – 18,583,753 |
| Length | 132 |
| Max. P | 0.898282 |

| Location | 18,583,621 – 18,583,736 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 83.42 |
| Mean single sequence MFE | -37.62 |
| Consensus MFE | -30.82 |
| Energy contribution | -30.82 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.594979 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
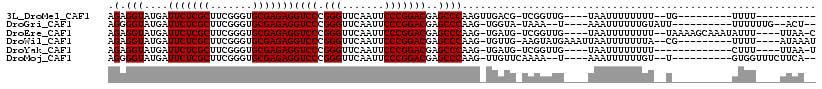
>3L_DroMel_CAF1 18583621 115 - 23771897 --CAACCGACGUC--AACUUGGGCUCGUCCGGGAAUUGAACCCGGGACCUCUCGCACCCGAAGCGAGAAUCAUACCUCUAGACCAACGAGCCUGCGGUACUGGGAGCGAUCAGAAUGAC --........(((--((((((((((((((((((.......)))))((..((((((.......)))))).))..............))))))))).))).((((......))))..)))) ( -38.30) >DroVir_CAF1 9159 111 - 1 --A--CAUUAACC--A-CUUGGGCUCGUCCGGGAAUUGAACCCGGGACCUCUCGCACCCGAAGCGAGAAUCAUACCCCUAGACCAACGAGCCAGUUGAAGGUGGAUCG-CCAUAUUGUA --.--......((--(-(((.((((((((((((.......)))))((..((((((.......)))))).))..............)))))))......))))))....-.......... ( -37.90) >DroPse_CAF1 20371 115 - 1 --UAAUUCACUUCCAC-CUUGGGCUCGUCCGGGAAUUGAACCCGGGACCUCUCGCACCCGAAGCGAGAAUCAUACCUCUAGACCAACGAGCCAGUUGAACAUGGAGAG-UCCUAUUGUG --...........(((-...((((((((((.((........)).)))).((((((.......))))))........((((...((((......))))....)))))))-)))....))) ( -35.20) >DroWil_CAF1 40632 115 - 1 UUCAUACUUCAAC--A-CUUGGGCUCGUCCGGGAAUUGAACCCGGGACCUCUCGCACCCGAAGCGAGAAUCAUACCUCUAGACCAACGAGCCGGUUGAAGGUUGAGCG-CCGCAUUGUC ((((..(((((((--.-....((((((((((((.......)))))((..((((((.......)))))).))..............))))))).)))))))..))))..-.......... ( -41.20) >DroAna_CAF1 17955 112 - 1 --AUUUCAAAUUA--G-CUUGGGCUCGUCCGGGAAUUGAACCCGGGACCUCUCGCACCCGAAGCGAGAAUCAUACCUCUAGACCAACGAGCCAGUUGAAGGUGGAGCGC-CGGAUUGU- --..(((...(((--(-((..((((((((((((.......)))))((..((((((.......)))))).))..............))))))))))))).((((...)))-))))....- ( -37.90) >DroPer_CAF1 21027 115 - 1 --UAAUUCACUUCCAC-CUUGGGCUCGUCCGGGAAUUGAACCCGGGACCUCUCGCACCCGAAGCGAGAAUCAUACCUCUAGACCAACGAGCCAGUUGAACAUGGAGAG-UCCUAUUGUG --...........(((-...((((((((((.((........)).)))).((((((.......))))))........((((...((((......))))....)))))))-)))....))) ( -35.20) >consensus __AAAUCCACUUC__A_CUUGGGCUCGUCCGGGAAUUGAACCCGGGACCUCUCGCACCCGAAGCGAGAAUCAUACCUCUAGACCAACGAGCCAGUUGAACGUGGAGCG_UCAUAUUGUC .....................((((((((((((.......)))))((..((((((.......)))))).))..............)))))))........................... (-30.82 = -30.82 + -0.00)

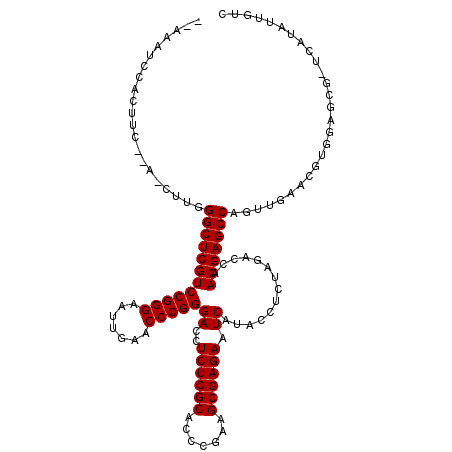
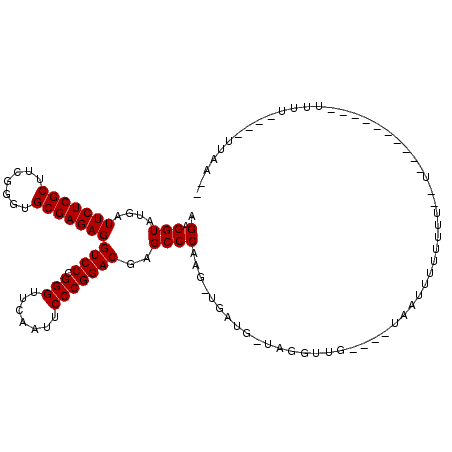
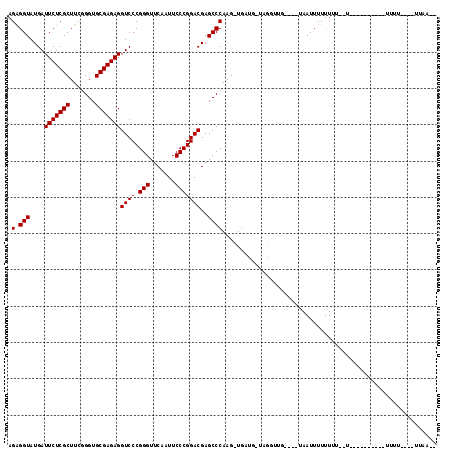
| Location | 18,583,661 – 18,583,753 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 78.43 |
| Mean single sequence MFE | -27.25 |
| Consensus MFE | -23.50 |
| Energy contribution | -23.50 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.85 |
| Structure conservation index | 0.86 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.521936 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18583661 92 + 23771897 AGAGGUAUGAUUCUCGCUUCGGGUGCGAGAGGUCCCGGGUUCAAUUCCCGGACGAGCCCAAGUUGACG-UCGGUUG----UAAUUUUUUUU--UG---------UUUU---------- .((.((....(((((((.......)))))))((((.(((.......)))))))............)).-)).....----...........--..---------....---------- ( -23.90) >DroGri_CAF1 9570 96 + 1 AGGGGUAUGAUUCUCGCUUCGGGUGCGAGAGGUCCCGGGUUCAAUUCCCGGACGAGCCCAAG-UGGUA-UAAA--U----AAAUUUUUUGUAUU----------UUUUUUG--ACU-- ..((((....(((((((.......)))))))((((.(((.......)))))))..))))(((-..(((-((((--.----......))))))).----------.)))...--...-- ( -29.90) >DroEre_CAF1 19364 105 + 1 AGAGGUAUGAUUCUCGCUUCGGGUGCGAGAGGUCCCGGGUUCAAUUCCCGGACGAGCCCAAG-UGAUG-UCGGUUG----UAAUUUUUUUU--UAAAAGCAAAUAUUU----UUAA-C (((((((((((..((((((.((((.(....)((((.(((.......)))))))..)))))))-))).)-))..(((----(..((((....--.))))))))))))))----))..-. ( -28.40) >DroWil_CAF1 40671 101 + 1 AGAGGUAUGAUUCUCGCUUCGGGUGCGAGAGGUCCCGGGUUCAAUUCCCGGACGAGCCCAAG-UGUUG-AAGUAUGAAAUUAAUUUUUUUA--CG---------UUUU----AUAAAU .(.(((....(((((((.......)))))))((((.(((.......)))))))..))))...-...((-(((..(((((.......)))))--..---------))))----)..... ( -26.80) >DroYak_CAF1 21255 94 + 1 AGAGGUAUGAUUCUCGCUUCGGGUGCGAGAGGUCCCGGGUUCAAUUCCCGGACGAGCCCAAG-UGAUG-UCGGUUG----UAAUUUUUUUU-------------CUUU----UUAA-U .......((((..((((((.((((.(....)((((.(((.......)))))))..)))))))-))).)-)))....----...........-------------....----....-. ( -25.70) >DroMoj_CAF1 10060 97 + 1 AGGGGUAUGAUUCUCGCUUCGGGUGCGAGAGGUCCCGGGUUCAAUUCCCGGACGAGCCCAAG-UUGUUCAAAA--U----AAAUUUUUUGU--U----------GUGGUUUCUUCA-- ..((((....(((((((.......)))))))((((.(((.......)))))))..))))(((-....(((.((--(----((.....))))--)----------.)))...)))..-- ( -28.80) >consensus AGAGGUAUGAUUCUCGCUUCGGGUGCGAGAGGUCCCGGGUUCAAUUCCCGGACGAGCCCAAG_UGAUG_UAGGUUG____UAAUUUUUUUU__U__________UUUU____UUAA__ .(.(((....(((((((.......)))))))((((.(((.......)))))))..))))........................................................... (-23.50 = -23.50 + 0.00)
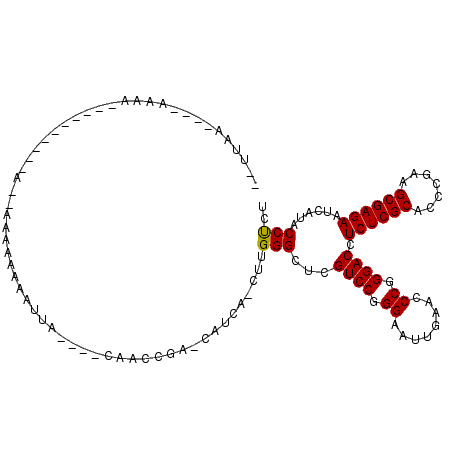
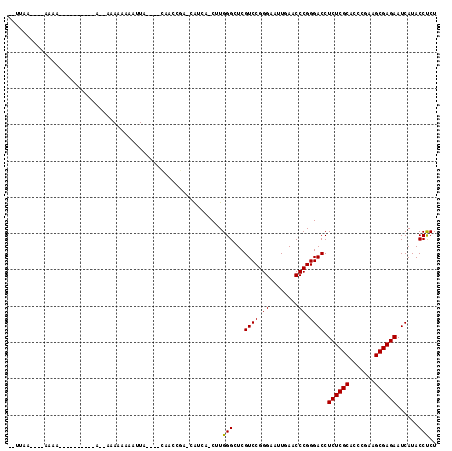
| Location | 18,583,661 – 18,583,753 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 78.43 |
| Mean single sequence MFE | -21.72 |
| Consensus MFE | -20.75 |
| Energy contribution | -20.53 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.898282 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18583661 92 - 23771897 ----------AAAA---------CA--AAAAAAAAUUA----CAACCGA-CGUCAACUUGGGCUCGUCCGGGAAUUGAACCCGGGACCUCUCGCACCCGAAGCGAGAAUCAUACCUCU ----------....---------..--...........----....(((-.(((......))))))((((((.......))))))...((((((.......))))))........... ( -22.20) >DroGri_CAF1 9570 96 - 1 --AGU--CAAAAAA----------AAUACAAAAAAUUU----A--UUUA-UACCA-CUUGGGCUCGUCCGGGAAUUGAACCCGGGACCUCUCGCACCCGAAGCGAGAAUCAUACCCCU --...--.......----------..............----.--....-.....-...(((...((((.((........)).)))).((((((.......)))))).......))). ( -22.20) >DroEre_CAF1 19364 105 - 1 G-UUAA----AAAUAUUUGCUUUUA--AAAAAAAAUUA----CAACCGA-CAUCA-CUUGGGCUCGUCCGGGAAUUGAACCCGGGACCUCUCGCACCCGAAGCGAGAAUCAUACCUCU .-((((----((........)))))--)..........----...((((-.....-.))))....((((.((........)).)))).((((((.......))))))........... ( -21.20) >DroWil_CAF1 40671 101 - 1 AUUUAU----AAAA---------CG--UAAAAAAAUUAAUUUCAUACUU-CAACA-CUUGGGCUCGUCCGGGAAUUGAACCCGGGACCUCUCGCACCCGAAGCGAGAAUCAUACCUCU ......----....---------.(--((....................-.....-...((.....((((((.......)))))).))((((((.......))))))....))).... ( -20.30) >DroYak_CAF1 21255 94 - 1 A-UUAA----AAAG-------------AAAAAAAAUUA----CAACCGA-CAUCA-CUUGGGCUCGUCCGGGAAUUGAACCCGGGACCUCUCGCACCCGAAGCGAGAAUCAUACCUCU .-....----....-------------...........----...((((-.....-.))))....((((.((........)).)))).((((((.......))))))........... ( -20.90) >DroMoj_CAF1 10060 97 - 1 --UGAAGAAACCAC----------A--ACAAAAAAUUU----A--UUUUGAACAA-CUUGGGCUCGUCCGGGAAUUGAACCCGGGACCUCUCGCACCCGAAGCGAGAAUCAUACCCCU --............----------.--.(((((.....----.--))))).....-...(((...((((.((........)).)))).((((((.......)))))).......))). ( -23.50) >consensus __UUAA____AAAA__________A__AAAAAAAAUUA____CAACCGA_CAUCA_CUUGGGCUCGUCCGGGAAUUGAACCCGGGACCUCUCGCACCCGAAGCGAGAAUCAUACCUCU ...........................................................(((...((((.((........)).)))).((((((.......))))))......))).. (-20.75 = -20.53 + -0.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:32:15 2006