
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 18,547,843 – 18,547,937 |
| Length | 94 |
| Max. P | 0.632520 |

| Location | 18,547,843 – 18,547,937 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 67.36 |
| Mean single sequence MFE | -37.41 |
| Consensus MFE | -12.66 |
| Energy contribution | -13.72 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.34 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.632520 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18547843 94 + 23771897 CGG-UCAGGGUGACGGUGAUUCUGGACAGUUGGUUUAAGCACCUGUCAACCCGUCACGAAGUG-U--------ACCCA--------------GUUACCAG-GACUGUGG-ACUGGUCAAA .(.-.(((.(((((((.(......(((((.((.......)).)))))..))))))))......-.--------.((((--------------(((.....-))))).))-.)))..)... ( -32.50) >DroSec_CAF1 171875 113 + 1 CGG-CCAGGGUGACGGUGAUUCUGUGCACUUGGUUUAAGCACCUGUCAACCAGUCACCCAGUG-CCCAGUACCACCCAGUUACCAACUG---GUUACCAG-GGCUGUGG-ACUGGUCAAA .((-((.(((((((((((((...((((...........))))..))).))).)))))))(((.-((((((.((..(((((.....))))---)......)-))))).))-)))))))... ( -44.30) >DroSim_CAF1 159656 109 + 1 CGG-CCAGGGUGACGGUGAUUCUGGGCACUUGGUUUAAGCACCUGUCAACCAGUCACCCAGUG-CCCAGUACCACCCAGUUACCA-------GUUACCAG-GGCUGUGG-ACUGGUCAAA .((-((((((((((((((...(((((((((.(((........(((.....)))..))).))))-)))))...))))..)))))).-------....(((.-.....)))-.))))))... ( -46.10) >DroEre_CAF1 168513 115 + 1 UAG-UCAGGGUGACGCUGAUUCUGGGCACUUGGCUUAAGCACCUGUCAUCCAGUCACCCAGUCACCCA--AUCACCCAGUUGCCAGUUGCCAGUUACCAG-GUCUGUGG-ACUGGUCAAA ...-...(((((((.......(((((.((...((....))....))..))))).......))))))).--.....((((((..(((..(((........)-)))))..)-)))))..... ( -36.14) >DroYak_CAF1 174226 97 + 1 UAG-CCAGGGUGACGGUGAUUCUGGGCAGUUGGCUUAAGCACCUGUCAUCCCGUCACCCAGCCAUCCA------C--------------CCAGUUACCAG-GGAUGUGG-ACUGGUCAAA ...-...(((((((((.(((....(((((...((....))..))))))))))))))))).((((((((------(--------------((........)-))...)))-).)))).... ( -40.20) >DroPer_CAF1 176964 80 + 1 UGGAGCGGUGGGACGGUGGGGCUCUGAGCCUGGUUUAAUCACCUGUCAGCCAGCUAC----------------------------------------GAACGGCUGAAACGCCAGUGGAA ...((((((..(((((((((((.....))))........)))).))).))).)))..----------------------------------------...(.((((......)))).).. ( -25.20) >consensus CGG_CCAGGGUGACGGUGAUUCUGGGCACUUGGUUUAAGCACCUGUCAACCAGUCACCCAGUG_CCCA_____ACCCA______________GUUACCAG_GGCUGUGG_ACUGGUCAAA .......(((((((((((((.....((...........))....)))).)).)))))))..........................................(((((......)))))... (-12.66 = -13.72 + 1.06)

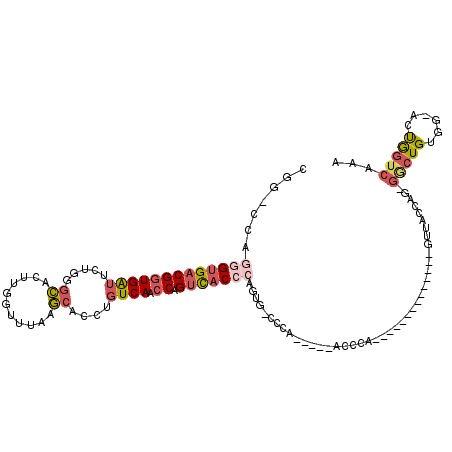
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:32:03 2006