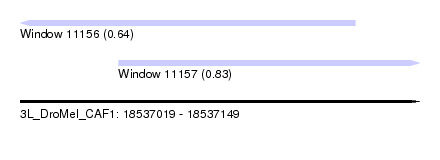
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 18,537,019 – 18,537,149 |
| Length | 130 |
| Max. P | 0.828218 |

| Location | 18,537,019 – 18,537,128 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 83.02 |
| Mean single sequence MFE | -43.85 |
| Consensus MFE | -28.16 |
| Energy contribution | -29.56 |
| Covariance contribution | 1.40 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.638965 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
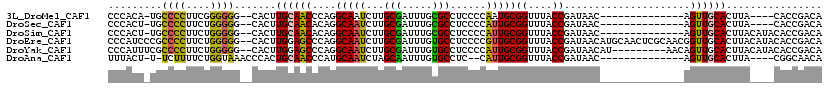
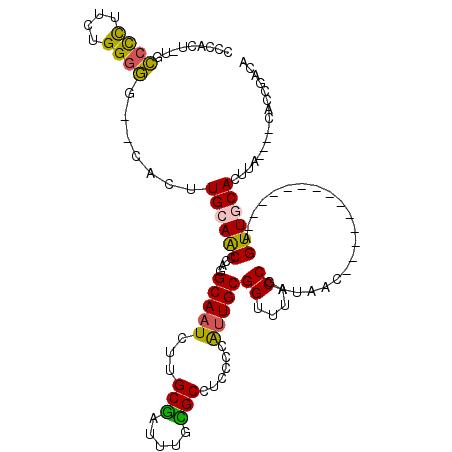
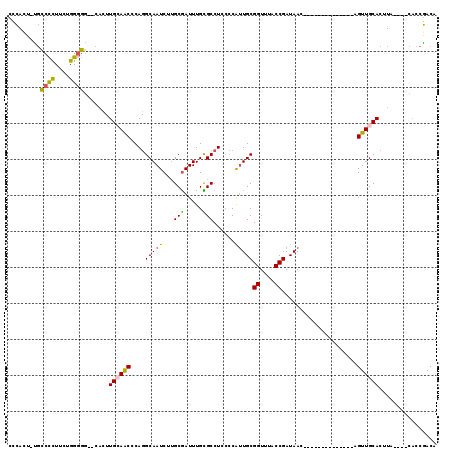
>3L_DroMel_CAF1 18537019 109 - 23771897 UAUCGGUAAACCGCAUUGGGGAGGCGCAAAUCGCAAGAUUGCCUGGGUUGCAAGUGCCCCCCGAAGGGGCA-UGUGGGCCAGCCACAUGCGCUGCAAUUGUG-UGCCUGCA ....((((...((((((((((.(((((.((((....))))((.......))..))))))))))).((.(((-(((((.....)))))))).)).....))))-)))).... ( -47.80) >DroPse_CAF1 166602 97 - 1 UAUCGGCAAACCGCAAUG--GAGGCACAAAUCGGCAG----CCAGGCAGCUGAUUGCCUC-GAAGGGGGCG-AA---GACAG---CACGCGGCGCACUUCUAUUGACUGCA .....(((.....(((((--((((..(.....)((.(----((..((.((((.(((((((-....))))))-).---..)))---)..))))))).)))))))))..))). ( -34.70) >DroSec_CAF1 161384 109 - 1 UAUCGGUAAACCGCAAUGGGGAGGCGCAAAUCGCAAGAUUGCCUGUGUUGCAAGUGCCCCCAGAAGGGGCA-AGUGGGGCAGCCACAUGCGCUGCAAUUGUG-UGCCUGCA ....((((...((((((...(.((((((((((....))))(.((((.(..(...((((((.....))))))-.)..).)))).)...)))))).).))))))-)))).... ( -45.00) >DroSim_CAF1 147391 109 - 1 UAUCGGUAAACCGCAAUGGGGAGGCGCAAAUCGCAAGAUUGCCUGUGUUGCAAGUGCCCCCAGAAGGGGCA-AGUGGGGCAGCCACAUGCGCUGCAAUUGUG-UGCCUGCA ....((((...((((((...(.((((((((((....))))(.((((.(..(...((((((.....))))))-.)..).)))).)...)))))).).))))))-)))).... ( -45.00) >DroEre_CAF1 158230 110 - 1 UAUCGGUAAACCGCAACGGGGAGGCACAAAUCGCAAGAUUGCCUGGGCUCCAAGUGCCCCCAGAAGGGGCGGGAUGGGGCAGCCACAUGUGCUGCAAUUGUG-UGCCUGCA ....((....))((....(((.(((((.((((....))))((....)).....))))))))...(((.(((.(((...((((((....).))))).))).))-).))))). ( -45.70) >DroYak_CAF1 163198 108 - 1 UAUCGGUAAACCGCAAUGGGGAGGCACAAAUCGCAAGAUUGCCUGGGCUCCAAGUGCCCCCAGAAGGGGCGAAAUGGGGCAGCCACAU--GCUGCAAUUGUG-UGCCUGCA ...(((....)))......(.(((((((((((....))))((....((((((..((((((.....))))))...))))))(((.....--))))).....))-))))).). ( -44.90) >consensus UAUCGGUAAACCGCAAUGGGGAGGCACAAAUCGCAAGAUUGCCUGGGUUCCAAGUGCCCCCAGAAGGGGCA_AAUGGGGCAGCCACAUGCGCUGCAAUUGUG_UGCCUGCA ....((....))((....(((.(((((.((((....))))((....)).....)))))))).....(((((.......(((((.......)))))........))))))). (-28.16 = -29.56 + 1.40)

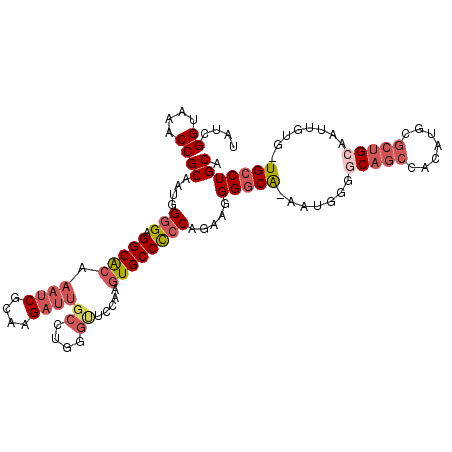
| Location | 18,537,051 – 18,537,149 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 80.15 |
| Mean single sequence MFE | -28.78 |
| Consensus MFE | -18.68 |
| Energy contribution | -18.55 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.828218 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18537051 98 + 23771897 CCCACA-UGCCCCUUCGGGGGG--CACUUGCAACCCAGGCAAUCUUGCGAUUUGCGCCUCCCCAAUGCGGUUUACCGAUAAC--------------AGUUGCACUUA----CACCGACA ......-((((((.....))))--))..((((((..((((((((....))))...))))........(((....))).....--------------.))))))....----........ ( -27.70) >DroSec_CAF1 161416 98 + 1 CCCACU-UGCCCCUUCUGGGGG--CACUUGCAACACAGGCAAUCUUGCGAUUUGCGCCUCCCCAUUGCGGUUUACCGAUAAC--------------AGUUGCACUUA----CACCGACA ......-((((((.....))))--))..((((((..((((((((....))))...))))........(((....))).....--------------.))))))....----........ ( -28.50) >DroSim_CAF1 147423 102 + 1 CCCACU-UGCCCCUUCUGGGGG--CACUUGCAACACAGGCAAUCUUGCGAUUUGCGCCUCCCCAUUGCGGUUUACCGAUAAC--------------AGUUGCACUUACAUACACCGACA ......-((((((.....))))--))..((((((..((((((((....))))...))))........(((....))).....--------------.))))))................ ( -28.50) >DroEre_CAF1 158262 117 + 1 CCCAUCCCGCCCCUUCUGGGGG--CACUUGGAGCCCAGGCAAUCUUGCGAUUUGUGCCUCCCCGUUGCGGUUUACCGAUAACAUGCAACUCGCAACGGUUGCACUUACAUACACCGACA .(((....(((((.....))))--)...))).......((((((((((((.(((((......)(((((((....))).))))..)))).)))))).))))))................. ( -37.50) >DroYak_CAF1 163228 108 + 1 CCCAUUUCGCCCCUUCUGGGGG--CACUUGGAGCCCAGGCAAUCUUGCGAUUUGUGCCUCCCCAUUGCGGUUUACCGAUAACAU---------AACAGUUGCACUUACAUACACCGACA ........(((((.....))))--)..((((.((....)).....(((((((.((............(((....))).......---------.)))))))))..........)))).. ( -26.35) >DroAna_CAF1 160597 97 + 1 UUUACU-U-UCUUUUCUGGUAAACCCACUGCAACCCAUGCAAUCUAGCAAUUUGUGCCUC--CAUUGCGGUUUACCGAUAAC--------------AGUUGCACUUA----CGGCAACA ......-.-.......(((((((((...((((.....)))).....(((((.........--.)))))))))))))).....--------------.(((((.....----..))))). ( -24.10) >consensus CCCACU_UGCCCCUUCUGGGGG__CACUUGCAACCCAGGCAAUCUUGCGAUUUGCGCCUCCCCAUUGCGGUUUACCGAUAAC______________AGUUGCACUUA____CACCGACA .........((((....)))).......((((((....(((((...(((.....)))......)))))((....)).....................))))))................ (-18.68 = -18.55 + -0.13)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:32:02 2006