
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 18,508,010 – 18,508,141 |
| Length | 131 |
| Max. P | 0.588159 |

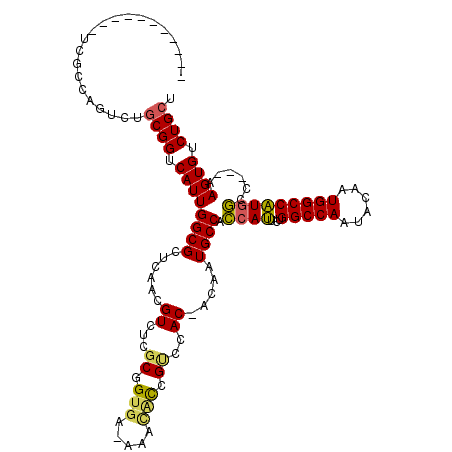
| Location | 18,508,010 – 18,508,113 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 82.95 |
| Mean single sequence MFE | -35.30 |
| Consensus MFE | -22.86 |
| Energy contribution | -22.62 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.588159 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
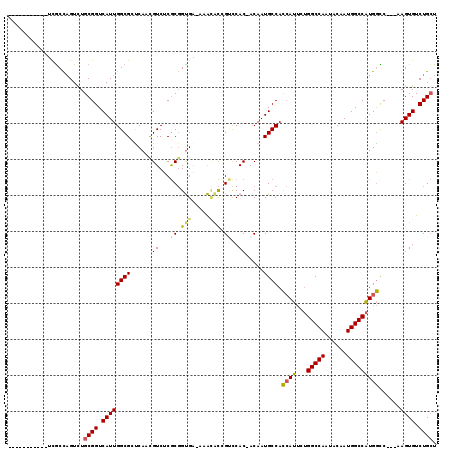
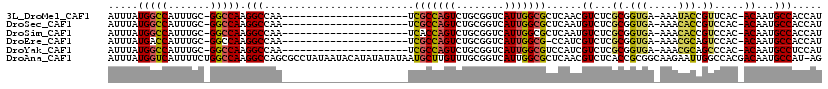
>3L_DroMel_CAF1 18508010 103 - 23771897 -----------UCGCCAGUCUGCGGUCAUUGGCGCUCAACGUCUCGCGGUGA-AAAUACCGUUCAC-ACAAUGCCACCAUUCUGGCCAAUACAAUGGCCAUGGCC---AAGUGUCUGCU -----------..((((((........))))))((..........((((((.-...))))))..((-((...((((......((((((......)))))))))).---..))))..)). ( -31.20) >DroSec_CAF1 132548 103 - 1 -----------UCGCCAGUCUGCGGUCAUUGGCGCUCAAUGUCUCGCGGUGA-AAACACCGUCCAC-ACAAUGCCACCAUUCUGGCCAAUACAAUGGCCAUGGUC---AAGUGUCUGCU -----------..........((((.((((((((.....(((...((((((.-...))))))..))-)...))))((((...((((((......)))))))))).---.)))).)))). ( -34.90) >DroSim_CAF1 117704 103 - 1 -----------UCACCAGUCUGCGGUCAUUGGCGCUCAAUGUCUCGCGGUGA-AAACACCGUCCAC-ACAAUGCCACCAUUUUGGCCAAUACAAUGGCCAUGGUC---AAGUGUCUGCU -----------..........((((.((((((((.....(((...((((((.-...))))))..))-)...))))((((...((((((......)))))))))).---.)))).)))). ( -34.90) >DroEre_CAF1 129691 102 - 1 -----------UCGCCAGUCUGCGGUCAUUGGCG-CCAUCGUCUCGCGGUGA-AAACGCAGUCCAC-ACAAUGCCACCAUUCUGGCCAAUACAAUGGCCAUGGCC---AAGUGUCUGCU -----------.(((((((........)))))))-.(((((.....))))).-....((((...((-((...((((......((((((......)))))))))).---..)))))))). ( -36.00) >DroYak_CAF1 133754 103 - 1 -----------UCGCCAGUCUGCGGUCAUUGGCGUCCAUCGUCUCGCGGUGA-AAACGCAGCCCAC-ACAAUGCCUCCAUUCUGGCCAAUACAAUGGCCAUGGCC---AAGUGUCUGCU -----------.(((((((........)))))))..(((((.....))))).-....((((...((-((...(((.......((((((......)))))).))).---..)))))))). ( -34.20) >DroAna_CAF1 128194 118 - 1 ACAUAUAUAUAAUGCUUGUUUGCGGUCAUUGGCGCUCAACGUCUCACCGCGGCAAGAAUUGGCCACGACAAUGCCAU-AGAAUGGCCAAUACAAUGGCCCUGACCGUGGAGUGUCUGGU .........((((.((((((.(((((....((((.....))))..))))))))))).))))((((.((((...(((.-.(...(((((......)))))....)..)))..)))))))) ( -40.60) >consensus ___________UCGCCAGUCUGCGGUCAUUGGCGCUCAACGUCUCGCGGUGA_AAACACCGUCCAC_ACAAUGCCACCAUUCUGGCCAAUACAAUGGCCAUGGCC___AAGUGUCUGCU .....................((((.((((((((......((...((.(((.....))).))..)).....)))).((((...(((((......)))))))))......)))).)))). (-22.86 = -22.62 + -0.24)

| Location | 18,508,046 – 18,508,141 |
|---|---|
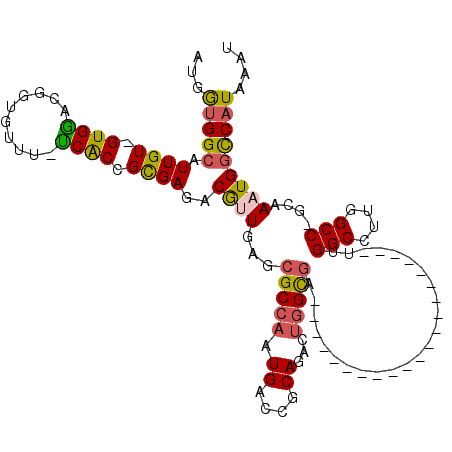
| Length | 95 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 80.91 |
| Mean single sequence MFE | -36.48 |
| Consensus MFE | -19.91 |
| Energy contribution | -19.88 |
| Covariance contribution | -0.02 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.586439 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18508046 95 + 23771897 AUGGUGGCAUUGU-GUGAACGGUAUUU-UCACCGCGAGACGUUGAGCGCCAAUGACCGCAGACUGGCGA---------------------UUGGCCUUGGCC-GCAAAUGGCCAUAAAU ...(((((....(-(((..((((....-..))))......((..((.((((((..(((.....)))..)---------------------)))))))..)))-)))....))))).... ( -33.50) >DroSec_CAF1 132584 95 + 1 AUGGUGGCAUUGU-GUGGACGGUGUUU-UCACCGCGAGACAUUGAGCGCCAAUGACCGCAGACUGGCGA---------------------UUGGCCUUGGCC-GCAAAUGGCCAUAAAU ...(((((....(-((((.(((((...-.))))).......(..((.((((((..(((.....)))..)---------------------)))))))..)))-)))....))))).... ( -36.50) >DroSim_CAF1 117740 95 + 1 AUGGUGGCAUUGU-GUGGACGGUGUUU-UCACCGCGAGACAUUGAGCGCCAAUGACCGCAGACUGGUGA---------------------UUGGCCUUGGCC-GCAAAUGGCCAUAAAU ...(((((....(-((((.(((((...-.))))).......(..((.((((((.((((.....)))).)---------------------)))))))..)))-)))....))))).... ( -40.30) >DroEre_CAF1 129727 94 + 1 AUGGUGGCAUUGU-GUGGACUGCGUUU-UCACCGCGAGACGAUGG-CGCCAAUGACCGCAGACUGGCGA---------------------UUGGCCUUGGCC-GCAAAUGGUCAUAAAU .(((((.((((((-(((((.......)-))))(....))))))).-)))))((((((((......))..---------------------..(((....)))-......)))))).... ( -34.60) >DroYak_CAF1 133790 95 + 1 AUGGAGGCAUUGU-GUGGGCUGCGUUU-UCACCGCGAGACGAUGGACGCCAAUGACCGCAGACUGGCGA---------------------UUGGCCUUGGCC-GCAAAUGGCCAUAAAU ...(((((....(-((.(((((((...-(((..(((..........)))...))).))))).)).))).---------------------...)))))((((-(....)))))...... ( -32.00) >DroAna_CAF1 128233 118 + 1 CU-AUGGCAUUGUCGUGGCCAAUUCUUGCCGCGGUGAGACGUUGAGCGCCAAUGACCGCAAACAAGCAUUAUAUAUAUGUAUUAUAGGCGCUGGCCUUGGCCAGAAAAUGACCAUAAAU .(-((((((((.((.(((((((.(((..(....)..))).(((.((((((.......((......))......((((.....))))))))))))).))))))))).)))).)))))... ( -42.00) >consensus AUGGUGGCAUUGU_GUGGACGGUGUUU_UCACCGCGAGACGUUGAGCGCCAAUGACCGCAGACUGGCGA_____________________UUGGCCUUGGCC_GCAAAUGGCCAUAAAU ...(((((.((((.((((..........)))).))))..((((...(((((.((....))...)))))........................(((....)))....))))))))).... (-19.91 = -19.88 + -0.02)


| Location | 18,508,046 – 18,508,141 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 80.91 |
| Mean single sequence MFE | -31.45 |
| Consensus MFE | -17.44 |
| Energy contribution | -17.50 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.540342 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18508046 95 - 23771897 AUUUAUGGCCAUUUGC-GGCCAAGGCCAA---------------------UCGCCAGUCUGCGGUCAUUGGCGCUCAACGUCUCGCGGUGA-AAAUACCGUUCAC-ACAAUGCCACCAU .....(((((......-))))).(((...---------------------.(((((((........)))))))......((...((((((.-...))))))..))-.....)))..... ( -27.90) >DroSec_CAF1 132584 95 - 1 AUUUAUGGCCAUUUGC-GGCCAAGGCCAA---------------------UCGCCAGUCUGCGGUCAUUGGCGCUCAAUGUCUCGCGGUGA-AAACACCGUCCAC-ACAAUGCCACCAU .....(((((......-)))))..(((((---------------------(.(((.......))).))))))((....(((...((((((.-...))))))..))-)....))...... ( -31.80) >DroSim_CAF1 117740 95 - 1 AUUUAUGGCCAUUUGC-GGCCAAGGCCAA---------------------UCACCAGUCUGCGGUCAUUGGCGCUCAAUGUCUCGCGGUGA-AAACACCGUCCAC-ACAAUGCCACCAU .....(((((......-)))))..(((((---------------------(.(((.......))).))))))((....(((...((((((.-...))))))..))-)....))...... ( -31.50) >DroEre_CAF1 129727 94 - 1 AUUUAUGACCAUUUGC-GGCCAAGGCCAA---------------------UCGCCAGUCUGCGGUCAUUGGCG-CCAUCGUCUCGCGGUGA-AAACGCAGUCCAC-ACAAUGCCACCAU ....((((((......-(((....)))..---------------------..((......))))))))(((((-.(((((.....))))).-.......((....-))..))))).... ( -25.20) >DroYak_CAF1 133790 95 - 1 AUUUAUGGCCAUUUGC-GGCCAAGGCCAA---------------------UCGCCAGUCUGCGGUCAUUGGCGUCCAUCGUCUCGCGGUGA-AAACGCAGCCCAC-ACAAUGCCUCCAU .....(((((......-)))))(((((((---------------------(.(((.......))).))))((((.(((((.....))))).-..)))).......-.....)))).... ( -29.40) >DroAna_CAF1 128233 118 - 1 AUUUAUGGUCAUUUUCUGGCCAAGGCCAGCGCCUAUAAUACAUAUAUAUAAUGCUUGUUUGCGGUCAUUGGCGCUCAACGUCUCACCGCGGCAAGAAUUGGCCACGACAAUGCCAU-AG ...(((((.((((.((((((((((((....))))...................((((((.(((((....((((.....))))..)))))))))))...)))))).)).))))))))-). ( -42.90) >consensus AUUUAUGGCCAUUUGC_GGCCAAGGCCAA_____________________UCGCCAGUCUGCGGUCAUUGGCGCUCAACGUCUCGCGGUGA_AAACACCGUCCAC_ACAAUGCCACCAU .....(((((.......))))).(((.........................(((((((........)))))))......((...((.(((.....))).)).....))...)))..... (-17.44 = -17.50 + 0.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:31:54 2006