
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 18,506,556 – 18,506,667 |
| Length | 111 |
| Max. P | 0.508941 |

| Location | 18,506,556 – 18,506,667 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.96 |
| Mean single sequence MFE | -28.75 |
| Consensus MFE | -17.58 |
| Energy contribution | -18.12 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.61 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.508941 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18506556 111 - 23771897 GUAGUGUCGGAAACAGGCCGCAAGCAUUUUCAUUUGCAUUCGGAAUGGAUUGCCGGACAUGGAAUAAAACCGUUUCCAAAAAA---------AAAAGUGAAUGCAAGUGGUCCAGAAACA ...((.(((....).((((((..((((..(((((((((.((......)).))).(((.((((.......)))).)))......---------..))))))))))..))))))..)).)). ( -31.90) >DroSec_CAF1 131131 113 - 1 GUAGUGUCGGAAACAGGCCGCAAGCAUUUUCAUUUGCAUUCGGAAUGGAUUGCCGGACAUGGAAUAAAACCGUUUCCAAAAAA-------AAGAAAGUGAACGCAAGUGGUCCAGAAACA ...((.(((....).((((((..((...((((((((((.((......)).))).(((.((((.......)))).)))......-------....))))))).))..))))))..)).)). ( -30.00) >DroSim_CAF1 116301 114 - 1 GUAGUGUCGGAAACAGGCCGCAAGCAUUUUCAUUUGCAUUCGGAAUGGAUUGCCGGACAUGGAAUAAAACCGUUUCCAAAAAA-----A-AAAAAAGUGAAUGCAAGUGGUCCAGAAACA ...((.(((....).((((((..((((..(((((((((.((......)).))).(((.((((.......)))).)))......-----.-....))))))))))..))))))..)).)). ( -31.90) >DroEre_CAF1 128229 109 - 1 GUAGUGUCGGAAACAGGCCGCAAGCAUUUUCAUUUGCAUUCGAAAUGGAUUGCCGGACAUGGAAUGAAACCGUUUCCAAAACG-------AA-AAAA---AUGCAAGUGGUCCAAAAACA ........(....).((((((..(((((((.....(((.((......)).))).(((.((((.......)))).)))......-------..-.)))---))))..))))))........ ( -28.30) >DroYak_CAF1 132194 112 - 1 GUAGUGUCGGAAACAGGCCGCAAGCAUUUUCAUUUGCAUUCGAAAUGGUUUGCCGGACAUGGAAUGAAACCGUAUCCAAAAAG-------AA-AAAAUGCAUGCAAGUGGUCCAAAAACA ........(....).((((((..((((...(((((...(((.....((....))(((.((((.......)))).))).....)-------))-.))))).))))..))))))........ ( -27.40) >DroPer_CAF1 133814 118 - 1 AAAGUGUCGUGAA-AAGCCGCAAGCAUUUUCAUUUGCAUGCAAAAUGUAGCGAUGGACAUGGGAUGAAAACGUUUCGGGGAAUUGGUAUCAA-AAAACGAAUUUAAAAACAACAACAGCA ...((((((((((-((..(....)..)))))))((((.(((.....)))))))..)))))(..(((....)))..)...(((((.((.....-...)).)))))................ ( -23.00) >consensus GUAGUGUCGGAAACAGGCCGCAAGCAUUUUCAUUUGCAUUCGAAAUGGAUUGCCGGACAUGGAAUAAAACCGUUUCCAAAAAA_______AA_AAAAUGAAUGCAAGUGGUCCAAAAACA ........(((.....(.(....))....((((((((((.(.....((....))(((.((((.......)))).))).....................).)))))))))))))....... (-17.58 = -18.12 + 0.53)

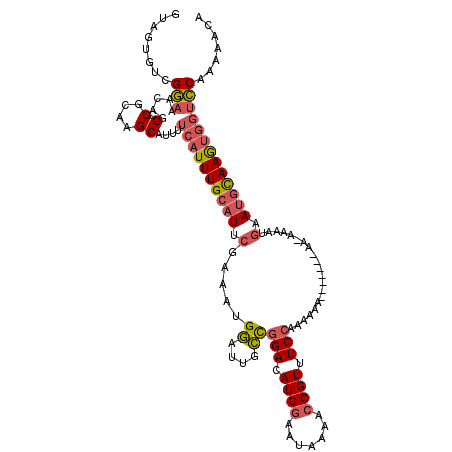
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:31:51 2006