
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 18,490,766 – 18,490,858 |
| Length | 92 |
| Max. P | 0.835876 |

| Location | 18,490,766 – 18,490,858 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 78.29 |
| Mean single sequence MFE | -31.62 |
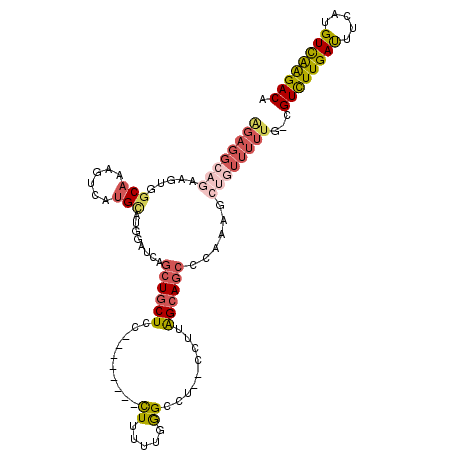
| Consensus MFE | -15.91 |
| Energy contribution | -17.14 |
| Covariance contribution | 1.23 |
| Combinations/Pair | 1.37 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.835876 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
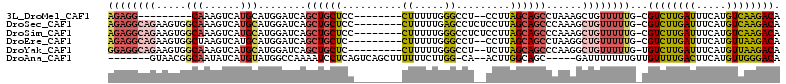
>3L_DroMel_CAF1 18490766 92 - 23771897 AGAGG---------CAAAGUCAUGCAUGGAUCAGCUGCUCC--------CUUUUUGGGCCU--CCUUAGCAGCCUAAAGCUGUUUUUG-CGUCUUGAUUUCAUGUCAAGACA ....(---------(((((.((.((.(((....((((((((--------(.....)))...--....)))))))))..)))).)))))-)((((((((.....)))))))). ( -30.31) >DroSec_CAF1 124429 103 - 1 AGAGGCAGAAGUGGCAAAGUCAUGCAUGGAUCAGCUGCUCC--------CUUUUUGAGCCUCUCCUUAGCAGCCCAAAGCUGUUUUUG-CGUCUUGAUUUCAUGUCAAGACA ....(((((((..((.......(((..(((..((..((((.--------......)))))).)))...))).......))..))))))-)((((((((.....)))))))). ( -33.64) >DroSim_CAF1 109507 103 - 1 AGAGGCAGAAGUGGCAAAGUCAUGCAUGGAUCAGCUGCUCC--------CUUUUUGGGCCUCUCCUUAGCAGCCCAAAGCUGUUUUUG-CGUCUUGAUUUCAUGUCAAGACA ....(((((((..((...(.....).(((....((((((((--------(.....))).........)))))))))..))..))))))-)((((((((.....)))))))). ( -36.80) >DroEre_CAF1 121581 100 - 1 AGAGGCAGAAGUGGCUAAGUCAUGCAUGGAUCAGCUGCUC---------CUUUUUGGGCCU--CCUUAGCAGCCUAAGGCUGUUUUUG-CGUCUUGAUUUCAUGUUAAGACA .(((((...((..(((...(((....)))...)))..))(---------(.....))))))--)...(((((((...)))))))....-.((((((((.....)))))))). ( -34.90) >DroYak_CAF1 125625 100 - 1 GGAGGCAGAAGUGGCAAAGUCAUGCAUGGAUCAGCUGCUC---------CUUUUUGGGCCU--UCUUAGCAGCCCAAGGCUGUUUUUG-UGUCUUGAUUUCAUGUUAAGACA ((((((...((..((....(((....)))....))..))(---------(.....))))))--))..(((((((...)))))))....-(((((((((.....))))))))) ( -33.60) >DroAna_CAF1 118828 97 - 1 -------GUAACGGCAAUAUCAUGUAUGGCCAAAAUCCUCAGUCAGCUUUUUUCUUGG-CA--ACUUGGCAGC-----GAUUUUUUUGUUGUUUUGACUUCAUGUUGGGACA -------.....(((.((((...)))).)))....(((.(((.(((((........))-).--.(..((((((-----((.....))))))))..)......)))))))).. ( -20.50) >consensus AGAGGCAGAAGUGGCAAAGUCAUGCAUGGAUCAGCUGCUCC________CUUUUUGGGCCU__CCUUAGCAGCCCAAAGCUGUUUUUG_CGUCUUGAUUUCAUGUCAAGACA ((((((((.....(((......)))........((((((..........((.....)).........))))))......))))))))...((((((((.....)))))))). (-15.91 = -17.14 + 1.23)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:31:49 2006