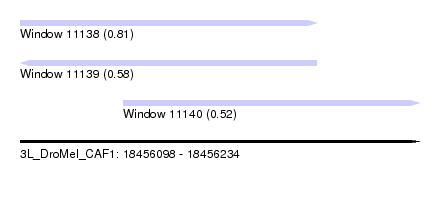
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 18,456,098 – 18,456,234 |
| Length | 136 |
| Max. P | 0.806144 |

| Location | 18,456,098 – 18,456,199 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 80.61 |
| Mean single sequence MFE | -34.15 |
| Consensus MFE | -17.58 |
| Energy contribution | -18.90 |
| Covariance contribution | 1.32 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.806144 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
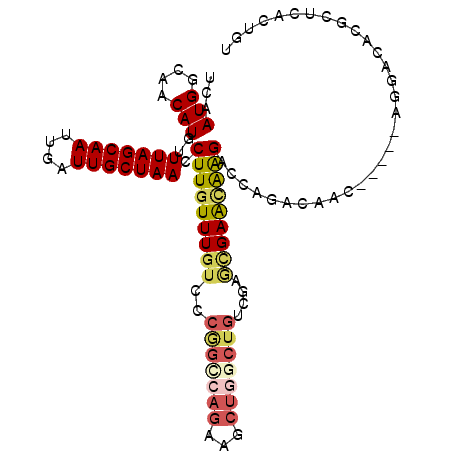
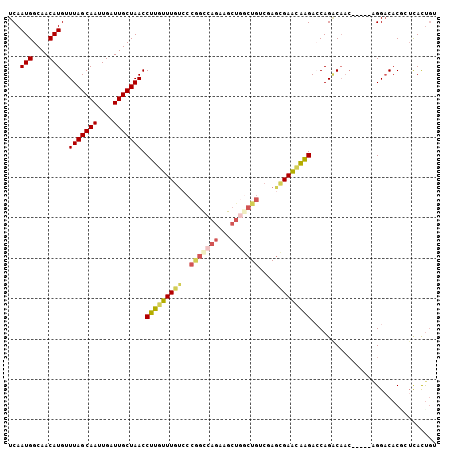
>3L_DroMel_CAF1 18456098 101 + 23771897 ACAGUGAGUGUGUCCU-----GUUGUCUGGUCUUGUUCGCUCGACAGCCAGCUUCUGACCGGGACAAAAAAGGUUAGCAAUCAAUUGCUAAACAUGUUGCCAUUGA .(((((((..((((((-----.((((((((((..(((.(((....))).)))....)))).))))))...)))(((((((....))))))))))..))..))))). ( -32.50) >DroSec_CAF1 96683 101 + 1 ACAGUGAGCGUGUCCU-----GUUGUCUGGUCUCGUUCACUGGACAGCCAGCUUCUGGCCGGGACAAACAAGGUUAGCAAUCAAUUGCUAAACAUGUUGCCAUUGA .(((((.((.((((((-----(.((((..((.......))..))))(((((...))))))))))))((((.(.(((((((....))))))).).))))))))))). ( -41.00) >DroSim_CAF1 87551 101 + 1 ACAGUGAGCGUGUCCU-----GUUGUCUGGUCUCGUUCACUGGCCAGCCAGCUUCUGGCCGGGACAAACAAGGUUAGCAAUCAAUUGCUAAACAUGUUGCCAUUGA .(((((.((.((((((-----(.....(((((.........)))))(((((...))))))))))))((((.(.(((((((....))))))).).))))))))))). ( -38.60) >DroEre_CAF1 94840 88 + 1 GCAGUGACAGUGUCCUUGC-----GCCUGGUCUUGCUCGCUCGACAGCCAG-------------CAAACAAGGUUAGCAAUCAAUUGCUAAACAUGUUGCCAUUGA .(((((.(((..(......-----((((.((.(((((.(((....))).))-------------))))).))))((((((....))))))...)..))).))))). ( -25.60) >DroYak_CAF1 97841 104 + 1 --AAUGACAGUGGCCGUUCCCGUUGUCUGCUCUUGUUCGCUCGACAGCCAGCUUCUGGCCGGGACAAACAAGGUUAGCAAUCAAUUGCUAAACAUGUUGCCAUUGA --.....(((((((.((.((((.((((.((........))..))))(((((...))))))))))).((((.(.(((((((....))))))).).))))))))))). ( -38.60) >DroAna_CAF1 92942 101 + 1 UCGAUAUCCGAGUCCC-----GUUGGCUGUUCCCAUUCGUUUGUCAGUUAGCUCCUGGCUGGGAUAAACAAGGUUAGCAAUCAAUUGCUAAACAUGUUGCCAUUGA (((((((((.((((..-----((((((((....((......)).))))))))....)))).)))))((((.(.(((((((....))))))).).))))....)))) ( -28.60) >consensus ACAGUGACCGUGUCCU_____GUUGUCUGGUCUCGUUCGCUCGACAGCCAGCUUCUGGCCGGGACAAACAAGGUUAGCAAUCAAUUGCUAAACAUGUUGCCAUUGA .(((((....((((((.....((((.(((...............))).))))........))))))((((.(.(((((((....))))))).).))))..))))). (-17.58 = -18.90 + 1.32)

| Location | 18,456,098 – 18,456,199 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 80.61 |
| Mean single sequence MFE | -29.73 |
| Consensus MFE | -18.72 |
| Energy contribution | -19.62 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.577607 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18456098 101 - 23771897 UCAAUGGCAACAUGUUUAGCAAUUGAUUGCUAACCUUUUUUGUCCCGGUCAGAAGCUGGCUGUCGAGCGAACAAGACCAGACAAC-----AGGACACACUCACUGU ...(((....)))..(((((((....)))))))(((...(((((..((((....(((.(....).)))......)))).))))).-----)))............. ( -24.30) >DroSec_CAF1 96683 101 - 1 UCAAUGGCAACAUGUUUAGCAAUUGAUUGCUAACCUUGUUUGUCCCGGCCAGAAGCUGGCUGUCCAGUGAACGAGACCAGACAAC-----AGGACACGCUCACUGU .....(((((((.(.(((((((....))))))).).)).))))).(((((((...)))))))..((((((.((.(.((.......-----.)).).)).)))))). ( -32.20) >DroSim_CAF1 87551 101 - 1 UCAAUGGCAACAUGUUUAGCAAUUGAUUGCUAACCUUGUUUGUCCCGGCCAGAAGCUGGCUGGCCAGUGAACGAGACCAGACAAC-----AGGACACGCUCACUGU .....(((((((.(.(((((((....))))))).).)).)))))((((((((...)))))))).((((((.((.(.((.......-----.)).).)).)))))). ( -35.30) >DroEre_CAF1 94840 88 - 1 UCAAUGGCAACAUGUUUAGCAAUUGAUUGCUAACCUUGUUUG-------------CUGGCUGUCGAGCGAGCAAGACCAGGC-----GCAAGGACACUGUCACUGC .((.(((((......(((((((....)))))))(((((((((-------------((.(((....))).)))))........-----))))))....))))).)). ( -27.40) >DroYak_CAF1 97841 104 - 1 UCAAUGGCAACAUGUUUAGCAAUUGAUUGCUAACCUUGUUUGUCCCGGCCAGAAGCUGGCUGUCGAGCGAACAAGAGCAGACAACGGGAACGGCCACUGUCAUU-- .((.((((((((.(.(((((((....))))))).).))))..((((((((((...)))))((((..((........)).)))).)))))...)))).)).....-- ( -36.90) >DroAna_CAF1 92942 101 - 1 UCAAUGGCAACAUGUUUAGCAAUUGAUUGCUAACCUUGUUUAUCCCAGCCAGGAGCUAACUGACAAACGAAUGGGAACAGCCAAC-----GGGACUCGGAUAUCGA ....((((((((.(.(((((((....))))))).).))))..(((((..(((.......))).(....)..)))))...))))..-----................ ( -22.30) >consensus UCAAUGGCAACAUGUUUAGCAAUUGAUUGCUAACCUUGUUUGUCCCGGCCAGAAGCUGGCUGUCGAGCGAACAAGACCAGACAAC_____AGGACACGCUCACUGU ...(((....)))..(((((((....))))))).(((((((((..(((((((...)))))))....)))))))))............................... (-18.72 = -19.62 + 0.89)
| Location | 18,456,133 – 18,456,234 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 86.49 |
| Mean single sequence MFE | -29.33 |
| Consensus MFE | -21.38 |
| Energy contribution | -22.85 |
| Covariance contribution | 1.47 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.73 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.517252 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
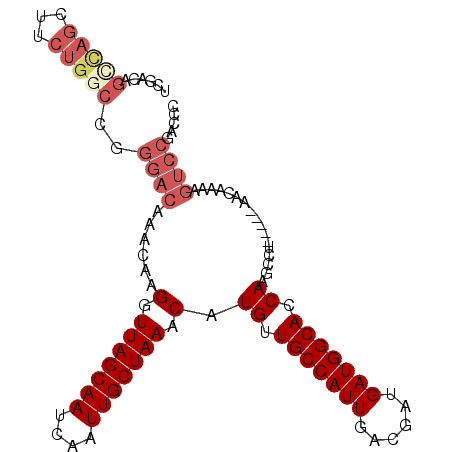
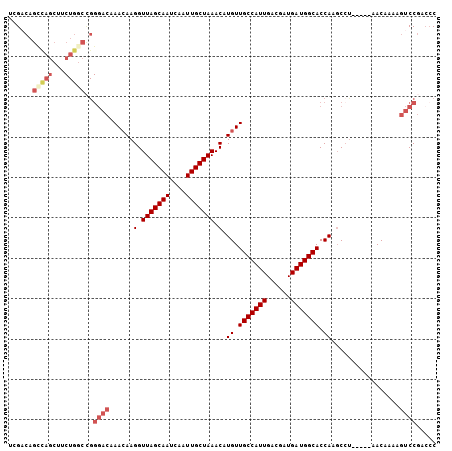
>3L_DroMel_CAF1 18456133 101 + 23771897 UCGACAGCCAGCUUCUGACCGGGACAAAAAAGGUUAGCAAUCAAUUGCUAAACAUGUUGCCAUUGACGAUGAUGGCACCAAGCCU-----AACAAAAGUCCGACCC ....(((.......)))....((((.....((((((((((....))))))....((.(((((((......))))))).)).))))-----.......))))..... ( -24.00) >DroSec_CAF1 96718 101 + 1 UGGACAGCCAGCUUCUGGCCGGGACAAACAAGGUUAGCAAUCAAUUGCUAAACAUGUUGCCAUUGACGAUGAUGGCACCAAGCCU-----AACAAAAGUCCGACCC .((((.(((((...))))).(((.(.((((.(.(((((((....))))))).).))))((((((......)))))).....))))-----.......))))..... ( -33.00) >DroSim_CAF1 87586 101 + 1 UGGCCAGCCAGCUUCUGGCCGGGACAAACAAGGUUAGCAAUCAAUUGCUAAACAUGUUGCCAUUGACGAUGAUGGCACCAAGCCU-----AACAAAAGUCCGACCC .((((((.......)))))).((((.((((.(.(((((((....))))))).).))))((((((......)))))).........-----.......))))..... ( -34.50) >DroEre_CAF1 94875 88 + 1 UCGACAGCCAG-------------CAAACAAGGUUAGCAAUCAAUUGCUAAACAUGUUGCCAUUGACGAUGAUGGCACCAUGCCU-----AACAAAAGUCCGACCC ..(((.(....-------------).....((((((((((....))))))).((((.(((((((......))))))).)))))))-----.......)))...... ( -23.40) >DroYak_CAF1 97879 101 + 1 UCGACAGCCAGCUUCUGGCCGGGACAAACAAGGUUAGCAAUCAAUUGCUAAACAUGUUGCCAUUGACGAUGAUGGCACCAUGCCU-----AACAAAAGUCCGACCC ....(.(((((...))))).)((((........(((((((....))))))).((((.(((((((......))))))).))))...-----.......))))..... ( -33.10) >DroAna_CAF1 92977 104 + 1 UUGUCAGUUAGCUCCUGGCUGGGAUAAACAAGGUUAGCAAUCAAUUGCUAAACAUGUUGCCAUUGACGAUGAUGGCACCACCCACUACUGCACCAUACUCCUU--C .((.((((.(((.....)))(((...((((.(.(((((((....))))))).).))))((((((......))))))....)))...))))))...........--. ( -28.00) >consensus UCGACAGCCAGCUUCUGGCCGGGACAAACAAGGUUAGCAAUCAAUUGCUAAACAUGUUGCCAUUGACGAUGAUGGCACCAAGCCU_____AACAAAAGUCCGACCC ......(((((...)))))..((((......(.(((((((....))))))).).((.(((((((......))))))).)).................))))..... (-21.38 = -22.85 + 1.47)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:31:45 2006