
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 18,441,678 – 18,441,774 |
| Length | 96 |
| Max. P | 0.986888 |

| Location | 18,441,678 – 18,441,774 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 72.17 |
| Mean single sequence MFE | -37.60 |
| Consensus MFE | -17.50 |
| Energy contribution | -17.83 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.43 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.47 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.803591 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
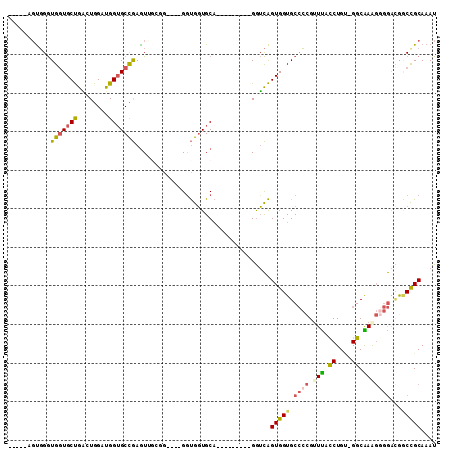
>3L_DroMel_CAF1 18441678 96 + 23771897 -----AGUGGGUGGUGCUGAGUGGAUGGAGCCGAGUUGGUGGAUGGAUGGUGCA---------GGUCAGUGGUGCCCCGUUUACCUGU-GGCAAAGAGGACGCCUGCAAAU -----.(..((((((.(.....).))...((((....(((((((((..((..(.---------........)..)))))))))))..)-)))........))))..).... ( -33.00) >DroSec_CAF1 82916 92 + 1 -----AGUGGGUGGUGCUGAGUGGAUGGUGCCGAGUUGCGG----GGUGGUGCA---------GGUCAGUGGUGCCCCGGUUACCUGU-GGCAAAGGGGACGACCGCAAAU -----.((((.(((..((........))..))).....(((----((((.(((.---------.....))).)))))))((..(((..-.....)))..))..)))).... ( -33.60) >DroSim_CAF1 75744 92 + 1 -----AGUGGGUGGUGCUGAGUGGAUGGUGCCGAGUUGCGG----GGUGGUGCA---------GGUCAGUGGUGCCCCGGUUACCUGU-GGCAAAGGGGACGACCGCAAAU -----.((((.(((..((........))..))).....(((----((((.(((.---------.....))).)))))))((..(((..-.....)))..))..)))).... ( -33.60) >DroEre_CAF1 80784 88 + 1 -----AGUGGGUGGUGCUGGCUGGGUGGUGCCGAAU----G----GGUGGUGCA---------GGUCAGUGGUGCCCCGUUUCUCUGU-GGCGAGGGGGGCAGCCGCAAAU -----.....((((((((((((..(((.((((....----.----)))).))).---------)))))))..((((((...((((...-...))))))))))))))).... ( -37.60) >DroYak_CAF1 83042 93 + 1 GGUUGAGUGGGUGGUGCUGACUGGCUGGUGCCGAGU----G----GGUGGUGCA---------GGUCAGUGGUGCCCCGUUCACCUGC-GGCGAAGGGGAUGGCCGCAAAU (((.(((((((..(..((.(((((((.(..(((...----.----..)))..).---------)))))))))..)))))))))))(((-(((.(......).))))))... ( -42.90) >DroAna_CAF1 68664 105 + 1 CACCACCCUGAUGCUGCCACCUGGCUGGUGCUAAGUGGCGU----GGCGCUGCCACAGAGAGGGAGUGGUGGUGCAAAGUUUACCUGGUGGCAAG--CAAAAGCCGCAAAU (((((((((....(((((((.((((....)))).)))))((----(((...)))))))...))).))))))................(((((...--.....))))).... ( -44.90) >consensus _____AGUGGGUGGUGCUGACUGGAUGGUGCCGAGUUGCGG____GGUGGUGCA_________GGUCAGUGGUGCCCCGUUUACCUGU_GGCAAAGGGGACGGCCGCAAAU ...........(((((((........)))))))...................................(((((.((((.(((.((....)).)))))))...))))).... (-17.50 = -17.83 + 0.34)


| Location | 18,441,678 – 18,441,774 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 72.17 |
| Mean single sequence MFE | -27.60 |
| Consensus MFE | -11.48 |
| Energy contribution | -12.07 |
| Covariance contribution | 0.59 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.42 |
| SVM decision value | 2.06 |
| SVM RNA-class probability | 0.986888 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
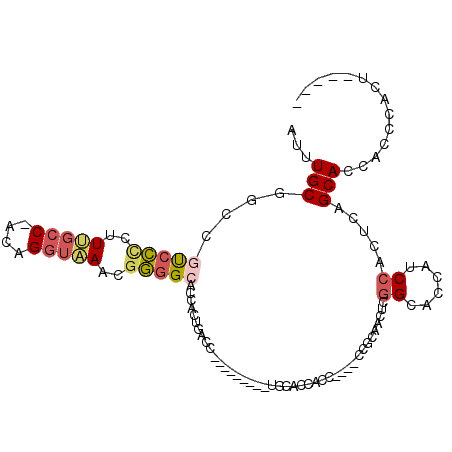
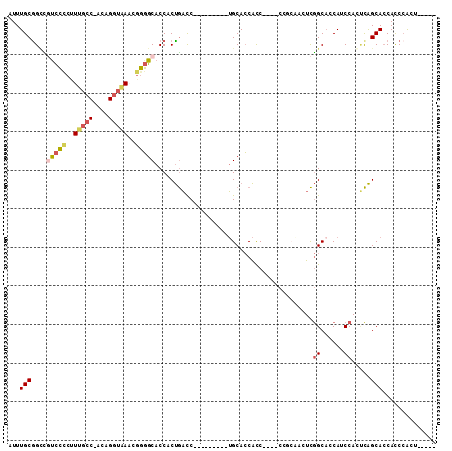
>3L_DroMel_CAF1 18441678 96 - 23771897 AUUUGCAGGCGUCCUCUUUGCC-ACAGGUAAACGGGGCACCACUGACC---------UGCACCAUCCAUCCACCAACUCGGCUCCAUCCACUCAGCACCACCCACU----- ...((((((((((((.((((((-...)))))).)))))......).))---------))))...................(((..........)))..........----- ( -19.80) >DroSec_CAF1 82916 92 - 1 AUUUGCGGUCGUCCCCUUUGCC-ACAGGUAACCGGGGCACCACUGACC---------UGCACCACC----CCGCAACUCGGCACCAUCCACUCAGCACCACCCACU----- ...((((((.(((((..(((((-...)))))..)))))))).((((..---------(((......----..)))..)))).............))).........----- ( -25.40) >DroSim_CAF1 75744 92 - 1 AUUUGCGGUCGUCCCCUUUGCC-ACAGGUAACCGGGGCACCACUGACC---------UGCACCACC----CCGCAACUCGGCACCAUCCACUCAGCACCACCCACU----- ...((((((.(((((..(((((-...)))))..)))))))).((((..---------(((......----..)))..)))).............))).........----- ( -25.40) >DroEre_CAF1 80784 88 - 1 AUUUGCGGCUGCCCCCCUCGCC-ACAGAGAAACGGGGCACCACUGACC---------UGCACCACC----C----AUUCGGCACCACCCAGCCAGCACCACCCACU----- ...(((((.((((((.(((...-...)))....)))))))).......---------.........----.----....(((........))).))).........----- ( -24.70) >DroYak_CAF1 83042 93 - 1 AUUUGCGGCCAUCCCCUUCGCC-GCAGGUGAACGGGGCACCACUGACC---------UGCACCACC----C----ACUCGGCACCAGCCAGUCAGCACCACCCACUCAACC ...((((((.(......).)))-)))((((...((....)).(((((.---------.((.....(----(----....)).....))..)))))))))............ ( -26.60) >DroAna_CAF1 68664 105 - 1 AUUUGCGGCUUUUG--CUUGCCACCAGGUAAACUUUGCACCACCACUCCCUCUCUGUGGCAGCGCC----ACGCCACUUAGCACCAGCCAGGUGGCAGCAUCAGGGUGGUG ...(((((..((((--((((....))))))))..))))).((((((((.......(((((...)))----))(((((((.((....)).))))))).......)))))))) ( -43.70) >consensus AUUUGCGGCCGUCCCCUUUGCC_ACAGGUAAACGGGGCACCACUGACC_________UGCACCACC____CCGCAACUCGGCACCAUCCACUCAGCACCACCCACU_____ ...(((....(((((..(((((....)))))..))))).........................................((......)).....))).............. (-11.48 = -12.07 + 0.59)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:31:41 2006