
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 18,434,025 – 18,434,137 |
| Length | 112 |
| Max. P | 0.825787 |

| Location | 18,434,025 – 18,434,115 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 79.38 |
| Mean single sequence MFE | -27.92 |
| Consensus MFE | -20.67 |
| Energy contribution | -21.70 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.17 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.648023 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
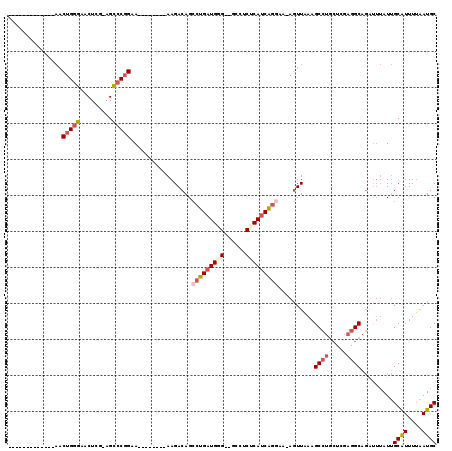
>3L_DroMel_CAF1 18434025 90 - 23771897 ----AAGACAGCCUGAUGGG--GCCUCUCAUCGGGAA-AGUUAAAGCCUGCUCGAGGCAGAUUUAUUGCAUUUUAAUGCGGGCUCUGGAUUGC---------------ACUC ----((..((((((((((((--....)))))))))..-......(((((((.....((((.....))))........))))))))))..))..---------------.... ( -28.62) >DroSec_CAF1 76339 90 - 1 ----AAGACAGCCUGAUGGG--GCCUCUCAUCAGGAA-AGUUAAAGCCUACUCGAGGCAGACUUAUUGCAUUUUAAUGCGGGCUCUGGAUUAC---------------ACUC ----((..((((((((((((--....))))))))).(-((((...((((.....)))).))))).((((((....))))))...)))..))..---------------.... ( -26.70) >DroSim_CAF1 70851 90 - 1 ----AAGACAGCCUGAUGGG--GCCUCUCAUCAGGAA-AGUUAAAGCCUGCUCGAGGCAGAUUUAUUGCAUUUUAAUGCGGGCUCUGGAUUGC---------------ACUC ----((..((((((((((((--....)))))))))..-......(((((((.....((((.....))))........))))))))))..))..---------------.... ( -29.32) >DroEre_CAF1 76103 88 - 1 ----AAGACAGCCUGAUG-G--GCCUCUCAUCAGG-A-AGUUUAAGCCUGCUCGAGGCAGAUUUAUUGCAUUUAAAUGCGGGCUCUGGAUUGC---------------GCCC ----.((((..(((((((-(--(...)))))))))-.-.))))..((((.....)))).........((((....))))((((.(......).---------------)))) ( -30.90) >DroYak_CAF1 77839 94 - 1 GGAAAAGACAGCCUGAUGCG--GCCUCUCAUCAGCAA-AGUUAAAGCCUGCUGGAGGCAGAUUUAUUGCAUUUUAAUGCGGGCUCUGGAUUGC---------------ACUC .((..(((..(((......)--)).)))..)).((((-..(((.((((((((((((((((.....)))).)))))..))))))).))).))))---------------.... ( -25.40) >DroAna_CAF1 63644 102 - 1 ----AAGACAGGAUGAUGAGCAACCACUCACCAGGAAAACUUAAAGCCUG------GCCCGUGAAUGGCGUUUUUAUGCGGGCUUCGAGCUGCCACAGGCAACUGCGAACUC ----....((((.((.((((......)))).)).............))))------((((((...............))))))((((((.((((...)))).)).))))... ( -26.60) >consensus ____AAGACAGCCUGAUGGG__GCCUCUCAUCAGGAA_AGUUAAAGCCUGCUCGAGGCAGAUUUAUUGCAUUUUAAUGCGGGCUCUGGAUUGC_______________ACUC ........((((((((((((......))))))))).........(((((((.....((((.....))))........))))))))))......................... (-20.67 = -21.70 + 1.03)


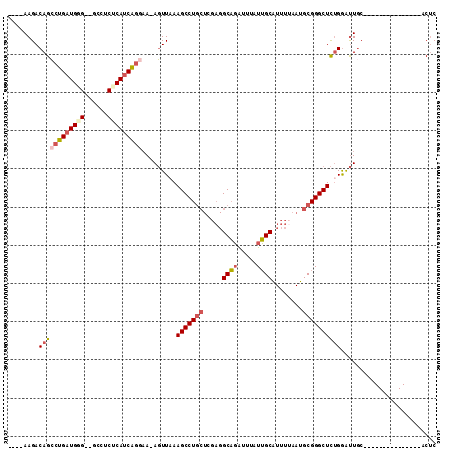
| Location | 18,434,043 – 18,434,137 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 77.37 |
| Mean single sequence MFE | -30.57 |
| Consensus MFE | -17.57 |
| Energy contribution | -18.65 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.825787 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
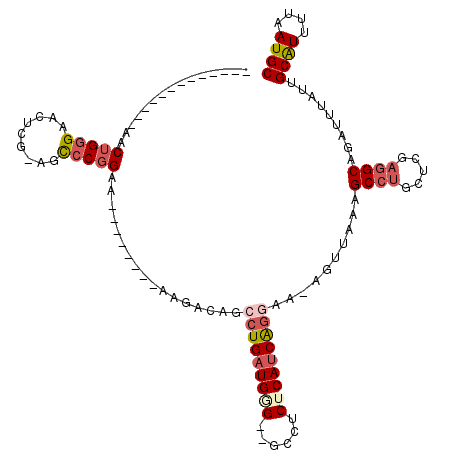
>3L_DroMel_CAF1 18434043 94 - 23771897 -------------AACUGAGAACUCG-AGCCCGGAA--------AAGACAGCCUGAUGGG--GCCUCUCAUCGGGAA-AGUUAAAGCCUGCUCGAGGCAGAUUUAUUGCAUUUUAAUGC -------------..(((....((((-(((..((..--------..(((..(((((((((--....)))))))))..-.)))....)).))))))).))).......((((....)))) ( -33.90) >DroSec_CAF1 76357 94 - 1 -------------AACUGGGAACUGG-AGCCCGGAA--------AAGACAGCCUGAUGGG--GCCUCUCAUCAGGAA-AGUUAAAGCCUACUCGAGGCAGACUUAUUGCAUUUUAAUGC -------------..(((((......-..)))))..--------.......(((((((((--....))))))))).(-((((...((((.....)))).)))))...((((....)))) ( -29.70) >DroSim_CAF1 70869 94 - 1 -------------AACUGGGAACUCG-AGCCCGGAA--------AAGACAGCCUGAUGGG--GCCUCUCAUCAGGAA-AGUUAAAGCCUGCUCGAGGCAGAUUUAUUGCAUUUUAAUGC -------------..(((....((((-(((..((..--------..(((..(((((((((--....)))))))))..-.)))....)).))))))).))).......((((....)))) ( -33.50) >DroEre_CAF1 76121 96 - 1 UGG---------CAACUGGGAGCUCC-UGCCCGGAA--------AAGACAGCCUGAUG-G--GCCUCUCAUCAGG-A-AGUUUAAGCCUGCUCGAGGCAGAUUUAUUGCAUUUAAAUGC ..(---------((((((((......-..)))))..--------.((((..(((((((-(--(...)))))))))-.-.))))..((((.....)))).......)))).......... ( -31.80) >DroYak_CAF1 77857 115 - 1 UCAGGAACUCGGCAACUGGGUACUUU-AGUCCGGAAAAGAGGAAAAGACAGCCUGAUGCG--GCCUCUCAUCAGCAA-AGUUAAAGCCUGCUGGAGGCAGAUUUAUUGCAUUUUAAUGC (((((.(((((....).)))).((((-..(((........)))))))....)))))....--(((((....(((((.-.((....)).)))))))))).........((((....)))) ( -29.60) >DroAna_CAF1 63677 90 - 1 ---------------CUGGGAACUCGAAGUCCUGAA--------AAGACAGGAUGAUGAGCAACCACUCACCAGGAAAACUUAAAGCCUG------GCCCGUGAAUGGCGUUUUUAUGC ---------------..(((..((((..((((((..--------....))))))..))))..........(((((...........))))------)))).......((((....)))) ( -24.90) >consensus _____________AACUGGGAACUCG_AGCCCGGAA________AAGACAGCCUGAUGGG__GCCUCUCAUCAGGAA_AGUUAAAGCCUGCUCGAGGCAGAUUUAUUGCAUUUUAAUGC ...............(((((.........))))).................(((((((((......)))))))))..........((((.....)))).........((((....)))) (-17.57 = -18.65 + 1.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:31:36 2006