
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 18,425,690 – 18,425,811 |
| Length | 121 |
| Max. P | 0.999967 |

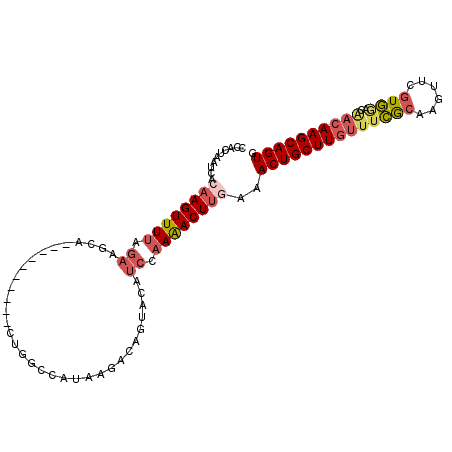
| Location | 18,425,690 – 18,425,784 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 69.53 |
| Mean single sequence MFE | -27.52 |
| Consensus MFE | -14.21 |
| Energy contribution | -15.96 |
| Covariance contribution | 1.75 |
| Combinations/Pair | 1.21 |
| Mean z-score | -3.69 |
| Structure conservation index | 0.52 |
| SVM decision value | 4.48 |
| SVM RNA-class probability | 0.999907 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18425690 94 + 23771897 CCACCACUCACAAGUUUUAGAGGCA----------CUGGUCAUAAGACAGCACCUCCAAAACUUAAAACUGCUUGUUUCGCAAGCUCGUGAACAACAAGCAGUG ...........(((((((.((((..----------(((.((....)))))..)))).)))))))...((((((((((((((......))))..)))))))))). ( -32.10) >DroSec_CAF1 61673 94 + 1 CUACUAAUCACAAGUUUUAGAAGCA----------CUGGCCAUAAGACAGUCCAUCCAAAACUUGAAACUGCUUGUUUCGCAAGUUCGUGGACGACAAGCAGUA ..........((((((((.((.(.(----------(((.........)))).).)).))))))))..((((((((((((((......))))..)))))))))). ( -29.70) >DroSim_CAF1 61061 94 + 1 CCACUAAUCACAAGUUUUAGAAGCA----------CUGGCCAUAAGACAGUACAUCCAAAACUUGAAACUGCUUGUUUCGCAAGUUCGUGGACGACAAGCAGUG ..........((((((((.((.(.(----------(((.........)))).).)).))))))))..((((((((((((((......))))..)))))))))). ( -29.70) >DroAna_CAF1 56880 99 + 1 UUAAUU-UUAUAAGUUUUAAAAGAGUAUGAUACAUUUAGCAAUGUUUCAAUAUCCCUCAGACUGUAAACUGCUU---UAAAAAGGUU-UUUAAAGCAAGCAGUU ......-...............(((..(((.(((((....))))).)))......))).((((((....(((((---(((((....)-))))))))).)))))) ( -18.60) >consensus CCACUAAUCACAAGUUUUAGAAGCA__________CUGGCCAUAAGACAGUACAUCCAAAACUUGAAACUGCUUGUUUCGCAAGUUCGUGGACAACAAGCAGUG ..........((((((((.((.................................)).))))))))..((((((((((((((......))))..)))))))))). (-14.21 = -15.96 + 1.75)


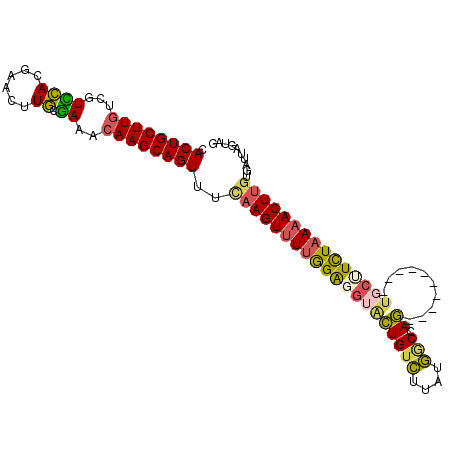
| Location | 18,425,690 – 18,425,784 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 69.53 |
| Mean single sequence MFE | -32.48 |
| Consensus MFE | -18.10 |
| Energy contribution | -18.48 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.38 |
| Mean z-score | -4.15 |
| Structure conservation index | 0.56 |
| SVM decision value | 5.00 |
| SVM RNA-class probability | 0.999967 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18425690 94 - 23771897 CACUGCUUGUUGUUCACGAGCUUGCGAAACAAGCAGUUUUAAGUUUUGGAGGUGCUGUCUUAUGACCAG----------UGCCUCUAAAACUUGUGAGUGGUGG (((..(((...........(((((.....))))).....(((((((((((((..(((((....)).)))----------..))))))))))))).)))..))). ( -41.50) >DroSec_CAF1 61673 94 - 1 UACUGCUUGUCGUCCACGAACUUGCGAAACAAGCAGUUUCAAGUUUUGGAUGGACUGUCUUAUGGCCAG----------UGCUUCUAAAACUUGUGAUUAGUAG .(((((((((..((((......)).)).)))))))))..(((((((((((.(.((((((....)).)))----------).).))))))))))).......... ( -31.70) >DroSim_CAF1 61061 94 - 1 CACUGCUUGUCGUCCACGAACUUGCGAAACAAGCAGUUUCAAGUUUUGGAUGUACUGUCUUAUGGCCAG----------UGCUUCUAAAACUUGUGAUUAGUGG ((((((((((..((((......)).)).))))))((((.(((((((((((.((((((((....)).)))----------))).))))))))))).)))))))). ( -35.20) >DroAna_CAF1 56880 99 - 1 AACUGCUUGCUUUAAA-AACCUUUUUA---AAGCAGUUUACAGUCUGAGGGAUAUUGAAACAUUGCUAAAUGUAUCAUACUCUUUUAAAACUUAUAA-AAUUAA .((((.((((((((((-(....)))))---))))))....)))).(((((((...(((.(((((....))))).)))...)))))))..........-...... ( -21.50) >consensus CACUGCUUGUCGUCCACGAACUUGCGAAACAAGCAGUUUCAAGUUUUGGAGGUACUGUCUUAUGGCCAG__________UGCUUCUAAAACUUGUGAUUAGUAG .((((((((...((((......)).))..))))))))..((((((((((((((((((((....))).))..........))))))))))))))).......... (-18.10 = -18.48 + 0.37)


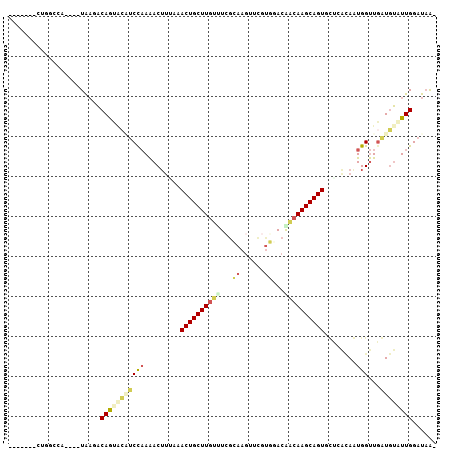
| Location | 18,425,715 – 18,425,811 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 67.88 |
| Mean single sequence MFE | -27.43 |
| Consensus MFE | -15.05 |
| Energy contribution | -15.91 |
| Covariance contribution | 0.87 |
| Combinations/Pair | 1.48 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.55 |
| SVM decision value | 2.14 |
| SVM RNA-class probability | 0.988915 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18425715 96 + 23771897 -------CUGGUCA----UAAGACAGCACCUCCAAAACUUAAAACUGCUUGUUUCGCAAGCUCGUGAACAACAAGCAGUGCUGAUAGUGGUUGGUGUAUUGGAUAAA -------...(((.----...))).(((((.(((....(((..((((((((((((((......))))..))))))))))..)))...)))..))))).......... ( -31.30) >DroSec_CAF1 61698 96 + 1 -------CUGGCCA----UAAGACAGUCCAUCCAAAACUUGAAACUGCUUGUUUCGCAAGUUCGUGGACGACAAGCAGUACUCACAGUGGUUGGUGUAUUGGAUAGA -------(((.(((----....(((..(((.(((.....(((.((((((((((((((......))))..))))))))))..)))...))).))))))..))).))). ( -30.00) >DroSim_CAF1 61086 96 + 1 -------CUGGCCA----UAAGACAGUACAUCCAAAACUUGAAACUGCUUGUUUCGCAAGUUCGUGGACGACAAGCAGUGCUCACAGUGGUUGGUGUGUUGGAUAGA -------.......----.....(((((((.((((.((((((.((((((((((((((......))))..))))))))))..))).)))..)))))))))))...... ( -31.20) >DroEre_CAF1 68914 93 + 1 -------CAGGCCGUUGGUAAGACAGUGCAUCUAAAACAUUGAACUGCUUGUCUGGCAAGA------ACGGCAAGCAGUCUUUAUAAUGGUUGAUGUAUUGGAUAG- -------...(((...)))....(((((((((.....(((((.((((((((.(((......------.))))))))))).....)))))...))))))))).....- ( -28.40) >DroYak_CAF1 70497 95 + 1 -------CAGGCCGUUUGUAAAACAGUGAGUCCAAAACUUUAAACUGCUUGUCUCGAAAGUUCGUUUACAACAAGCAGU----GCAAUGGUUGAUGUAUUGGAUAA- -------...((.(((.....))).))..((((((((((....(((((((((..(((....)))......)))))))))----.....))))......))))))..- ( -20.70) >DroAna_CAF1 56907 97 + 1 GAUACAUUUAGCAA----UGUUUCAAUAUCCCUCAGACUGUAAACUGCUU---UAAAAAGGUU-UUUAAAGCAAGCAGUUUGUA-AAUGGCUUAUAACUUGGCUUA- ((.(((((....))----))).)).........((((((((....(((((---(((((....)-))))))))).))))))))..-...((((........))))..- ( -23.00) >consensus _______CUGGCCA____UAAGACAGUACAUCCAAAACUUUAAACUGCUUGUUUCGCAAGUUCGUGGACAACAAGCAGUGCUCACAAUGGUUGAUGUAUUGGAUAA_ .......................(((((((((((.........((((((((((...((......))...))))))))))........)))...))))))))...... (-15.05 = -15.91 + 0.87)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:31:31 2006