
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 18,414,699 – 18,414,823 |
| Length | 124 |
| Max. P | 0.939748 |

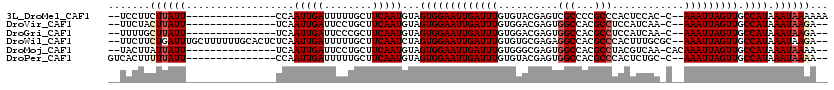
| Location | 18,414,699 – 18,414,799 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.52 |
| Mean single sequence MFE | -25.55 |
| Consensus MFE | -12.93 |
| Energy contribution | -13.43 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.51 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18414699 100 + 23771897 --UCCUUCUUAUU---------------CCAAUUGAUUUUUGCUUCAAUGUAGUGGAAUUGAUUUGUGUACGAGUCGCCCCGCCCACUCCAC-C--AAAUUAGUUGCCAUAAAUAAAAAA --......((((.---------------.((((((((((.(((......)))(((((...((((((....))))))((...))....)))))-.--))))))))))..))))........ ( -18.60) >DroVir_CAF1 132566 98 + 1 --UUCUACUUAUU---------------UCAAUUGAUUCCUGCUUCAAUGUAGUGGAAUUGAUUUGUGGACGAGUGGCCACGCCUCCAUCAA-C--AAAUUAGUUGCCAUAAAUAAGA-- --.....((((((---------------(..(((((........)))))...(((((((((((((((.((.(.(.(((...))).)).)).)-)--))))))))).))))))))))).-- ( -25.80) >DroGri_CAF1 198466 98 + 1 --UUUUGCUUAUU---------------UCAAUUGAUUCCCGCUUCAAUGUAGUGGAAUUGAUUUGUGGACGAGUGGCCACGCCUCCAUCAA-C--AAAUUAGUUGCCAUAAAUAAGA-- --.....((((((---------------(..(((((........)))))...(((((((((((((((.((.(.(.(((...))).)).)).)-)--))))))))).))))))))))).-- ( -25.50) >DroWil_CAF1 474413 114 + 1 --UUCUUCUGAUUUGCUUUUUUGCACUCUCAAUUGAUUUUUGCUUCAAUCUAGUGGAAUUGAUUUGUGUGCGAGAGGCCACGCCCACUUUGCGC--AAAUUAGUUGCCAUAAAUAAGA-- --.....(((((((((..((((((((.(..(((..(((.(..((.......))..))))..))).).))))))))(((...)))........))--)))))))...............-- ( -28.30) >DroMoj_CAF1 174662 100 + 1 --UACUUAUUAUU---------------UCAAUUGAUUCCUGCUUCAAUGUAGUGGAAUUGAUUUGUGGGCGAGUGGCCACGCCUACGUCAA-CACAAAUUAGUUGCCAUAAAUAAAA-- --......(((((---------------(..(((((........)))))...((((((((((((((((((((...(((...)))..))))..-)))))))))))).))))))))))..-- ( -26.80) >DroPer_CAF1 54506 100 + 1 GUCACUUUUUAUU---------------CCAAUUGAUUUUUGCUUCAAUGUAGUGGAAUUGAUUUGUGUACGAGUGGCCACGCCCACUCUGC-C--AAAUUAGUUGCCAUAAAUAAAA-- ......(((((((---------------...(((((........)))))...((((((((((((((.(((.((((((......)))))))))-)--))))))))).)))).)))))))-- ( -28.30) >consensus __UUCUUCUUAUU_______________UCAAUUGAUUCCUGCUUCAAUGUAGUGGAAUUGAUUUGUGGACGAGUGGCCACGCCCACUUCAA_C__AAAUUAGUUGCCAUAAAUAAAA__ .......((((((..................(((((........)))))...(((((((((((((..........(((...)))............))))))))).)))).))))))... (-12.93 = -13.43 + 0.50)
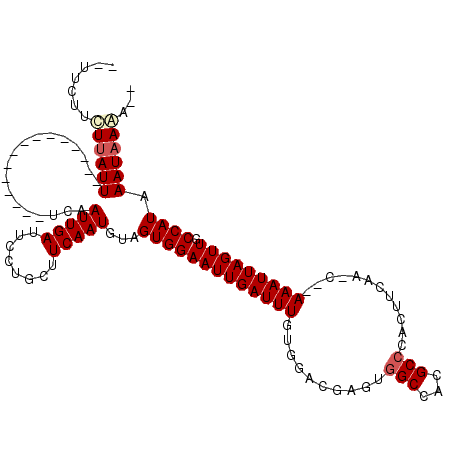

| Location | 18,414,722 – 18,414,823 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 79.62 |
| Mean single sequence MFE | -21.82 |
| Consensus MFE | -16.57 |
| Energy contribution | -16.90 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.939748 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18414722 101 + 23771897 UGCUUCAAUGUAGUGGAAUUGAUUUGUGUACGAGUCGCCCCGCCCACUCCAC-CAAAUUAGUUGCCAUAAAUAAAAAAAAAA-A-GAAAGGAAUCCAAACCAGA------- (((......)))((((((((((((((.((..((((.((...))..)))).))-)))))))))).))))..............-.-....((........))...------- ( -17.00) >DroVir_CAF1 132589 86 + 1 UGCUUCAAUGUAGUGGAAUUGAUUUGUGGACGAGUGGCCACGCCUCCAUCAA-CAAAUUAGUUGCCAUAAAUAAGA----AAAA-AAAUGGC------------------- (((......)))(((((((((((((((.((.(.(.(((...))).)).)).)-)))))))))).))))........----....-.......------------------- ( -19.70) >DroPse_CAF1 55298 85 + 1 UGCUUCAAUGUAGUGGAAUUGAUUUGUGUACGAGUGGCCACGCCCACUCUGC-CAAAUUAGUUGCCAUAAAUAAAA----A--A--AAAGGAA----------A------- (((......)))((((((((((((((.(((.((((((......)))))))))-)))))))))).))))........----.--.--.......----------.------- ( -25.20) >DroWil_CAF1 474451 85 + 1 UGCUUCAAUCUAGUGGAAUUGAUUUGUGUGCGAGAGGCCACGCCCACUUUGCGCAAAUUAGUUGCCAUAAAUAAGA----AA-A-GAAAAC-------------------- ............((((((((((((((((((.(((.(((...)))..))))))))))))))))).))))........----..-.-......-------------------- ( -21.20) >DroAna_CAF1 46099 105 + 1 UGCUUCAAUGUAGUGGAAUUGAUUUGUGUACGAGUGGCCACGCCCACUCCAC-CAAAUUAGUUGCCAUAAAUAAAA----AA-AUAAAAGAAAACCAAAACUGAAAACUGA (((......)))((((((((((((((.((..((((((......)))))).))-)))))))))).))))........----..-............................ ( -22.60) >DroPer_CAF1 54531 86 + 1 UGCUUCAAUGUAGUGGAAUUGAUUUGUGUACGAGUGGCCACGCCCACUCUGC-CAAAUUAGUUGCCAUAAAUAAAA----AA-A--AAAGGAA----------A------- (((......)))((((((((((((((.(((.((((((......)))))))))-)))))))))).))))........----..-.--.......----------.------- ( -25.20) >consensus UGCUUCAAUGUAGUGGAAUUGAUUUGUGUACGAGUGGCCACGCCCACUCCAC_CAAAUUAGUUGCCAUAAAUAAAA____AA_A__AAAGGAA__________A_______ ............((((((((((((((.....((((((......))))))....)))))))))).))))........................................... (-16.57 = -16.90 + 0.33)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:31:28 2006