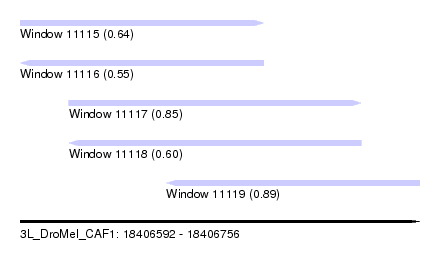
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 18,406,592 – 18,406,756 |
| Length | 164 |
| Max. P | 0.894682 |

| Location | 18,406,592 – 18,406,692 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.49 |
| Mean single sequence MFE | -21.28 |
| Consensus MFE | -14.86 |
| Energy contribution | -15.20 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.635088 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18406592 100 + 23771897 --------CAUUCU------------CAAAGGCAAAACAAAGAAAAGAAUUGUUUUCGCUUAAAAGAACAUUCAAAUGGAAAUGUUCUGCUUGGCUCAACACAAACAACAAGUGCAGGCG --------......------------...(((((((((((.........))))))).))))...((((((((........))))))))((((((((..............))).))))). ( -19.04) >DroPse_CAF1 45293 96 + 1 AG--GAGACAUUCU------------CAAAAGCAAAACAAAGAAGA-AAUUGUUUUCGCUUAAAAGAACAUUCAAAUGGAAAUGUUCUGCUUGGCUCAACACAAACAACAA--------- ..--(((.((..(.------------...(((((((((((......-..))))))).))))...((((((((........)))))))))..)).)))..............--------- ( -18.30) >DroEre_CAF1 50360 104 + 1 ----AAGGC------------AAAGGCAAAGGCAAAACAAAGAAAAGAAUUGUUUUCGCUUAAAAGAACAUUCAAAUGGAAAUGUUCUGCUUGGCUCAACACAAACAACCAGUGCAGGCG ----...((------------....(((..((.........(((((......)))))(((....((((((((........))))))))....))).............))..)))..)). ( -21.80) >DroWil_CAF1 463200 106 + 1 AGGCAAAAAAUUCU------------CAAAAGCAAAACAAAGAAGA-AAUUGUUUUCGCUUAAAAGAACAUUCAAAUGGAAAUGUUCUGCUUGGCUCAACACAAACAACAAGUGCAGGC- ((((..........------------...(((((((((((......-..))))))).))))...((((((((........)))))))))))).(((...(((.........)))..)))- ( -19.90) >DroYak_CAF1 51339 116 + 1 ----AAGGCAUUCUCCAGAGCAAAAGCAAGAGCAAAACAAAGAAAAGAAUUGUUUUCGCUUAAAAGAACAUUCAAAUGGAAAUGUUCUGCUUGGCUCAACACAAACAACAAGUGCAGGCG ----...((........((((..((((..(((((((((((.........))))))).))))...((((((((........)))))))))))).))))..(((.........)))...)). ( -27.50) >DroAna_CAF1 39325 104 + 1 ----AAGGGAUUCU------------CGAAAGCAAAAUAAAGAAAGGAAUUGUUUUCGCUUAAAAGAACAUUCAAAUGGAAAUGUUCUGCUUGGCUCAACACAAACAACGAAUGCAGGCG ----...(.((((.------------.((((((((..............))))))))(((....((((((((........))))))))....)))..............)))).)..... ( -21.14) >consensus ____AAGGCAUUCU____________CAAAAGCAAAACAAAGAAAAGAAUUGUUUUCGCUUAAAAGAACAUUCAAAUGGAAAUGUUCUGCUUGGCUCAACACAAACAACAAGUGCAGGCG .............................(((((((((((.........))))))).))))...((((((((........))))))))(((((.....................))))). (-14.86 = -15.20 + 0.34)

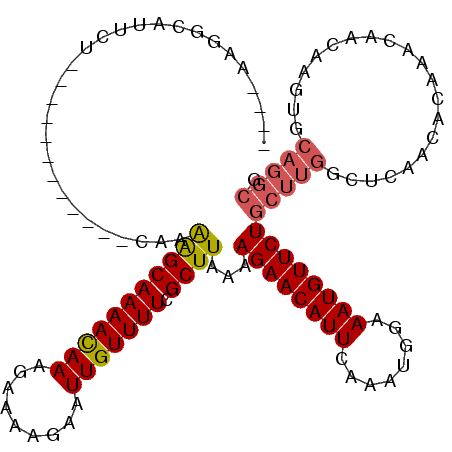
| Location | 18,406,592 – 18,406,692 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.49 |
| Mean single sequence MFE | -22.46 |
| Consensus MFE | -16.38 |
| Energy contribution | -16.55 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.554646 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18406592 100 - 23771897 CGCCUGCACUUGUUGUUUGUGUUGAGCCAAGCAGAACAUUUCCAUUUGAAUGUUCUUUUAAGCGAAAACAAUUCUUUUCUUUGUUUUGCCUUUG------------AGAAUG-------- .(((.((((.........)))).).))......((((((((......))))))))((((((((.(((((((.........)))))))))..)))------------)))...-------- ( -18.70) >DroPse_CAF1 45293 96 - 1 ---------UUGUUGUUUGUGUUGAGCCAAGCAGAACAUUUCCAUUUGAAUGUUCUUUUAAGCGAAAACAAUU-UCUUCUUUGUUUUGCUUUUG------------AGAAUGUCUC--CU ---------...(((((((.(.....)))))))).............((((((((((..((((.(((((((..-......)))))))))))..)------------))))))).))--.. ( -21.90) >DroEre_CAF1 50360 104 - 1 CGCCUGCACUGGUUGUUUGUGUUGAGCCAAGCAGAACAUUUCCAUUUGAAUGUUCUUUUAAGCGAAAACAAUUCUUUUCUUUGUUUUGCCUUUGCCUUU------------GCCUU---- .....(((..(((((((((.(.....)))))))((((((((......))))))))......((.(((((((.........)))))))))....)))..)------------))...---- ( -23.00) >DroWil_CAF1 463200 106 - 1 -GCCUGCACUUGUUGUUUGUGUUGAGCCAAGCAGAACAUUUCCAUUUGAAUGUUCUUUUAAGCGAAAACAAUU-UCUUCUUUGUUUUGCUUUUG------------AGAAUUUUUUGCCU -(((.((((.........)))).).))...(((((((((((......)))))(((((..((((.(((((((..-......)))))))))))..)------------))))..)))))).. ( -25.00) >DroYak_CAF1 51339 116 - 1 CGCCUGCACUUGUUGUUUGUGUUGAGCCAAGCAGAACAUUUCCAUUUGAAUGUUCUUUUAAGCGAAAACAAUUCUUUUCUUUGUUUUGCUCUUGCUUUUGCUCUGGAGAAUGCCUU---- ..((.((((.........)))).((((.(((((((((((((......)))))))).....(((.(((((((.........))))))))))..)))))..)))).))..........---- ( -27.10) >DroAna_CAF1 39325 104 - 1 CGCCUGCAUUCGUUGUUUGUGUUGAGCCAAGCAGAACAUUUCCAUUUGAAUGUUCUUUUAAGCGAAAACAAUUCCUUUCUUUAUUUUGCUUUCG------------AGAAUCCCUU---- .....(((((((.((...(((((..((...))..)))))...))..)))))))((((..((((((((................))))))))..)------------))).......---- ( -19.09) >consensus CGCCUGCACUUGUUGUUUGUGUUGAGCCAAGCAGAACAUUUCCAUUUGAAUGUUCUUUUAAGCGAAAACAAUUCUUUUCUUUGUUUUGCUUUUG____________AGAAUGCCUU____ .............((((((.(.....)))))))((((((((......))))))))......((.(((((((.........)))))))))............................... (-16.38 = -16.55 + 0.17)

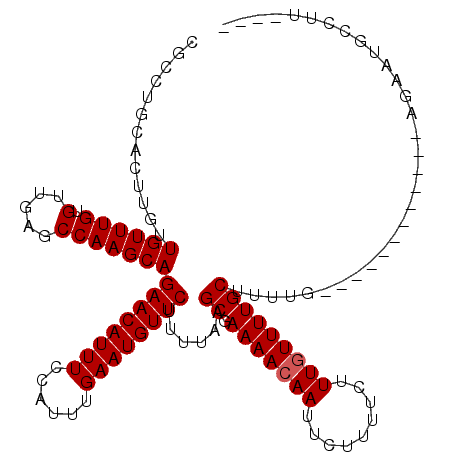
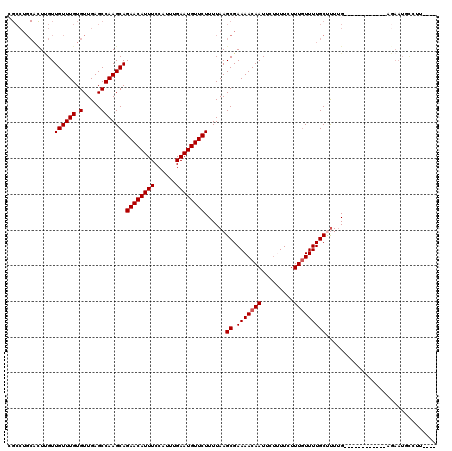
| Location | 18,406,612 – 18,406,732 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.19 |
| Mean single sequence MFE | -23.08 |
| Consensus MFE | -20.43 |
| Energy contribution | -21.10 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.851190 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18406612 120 + 23771897 AGAAAAGAAUUGUUUUCGCUUAAAAGAACAUUCAAAUGGAAAUGUUCUGCUUGGCUCAACACAAACAACAAGUGCAGGCGCACACACAAACUGCAUUUAUUUAGUUAGCAAACAAAAUAG .........((((((.((((....((((((((........))))))))....)))..............((((((((.............)))))))).........).))))))..... ( -23.92) >DroVir_CAF1 122606 115 + 1 AGAAGA-AAUUGUUUUCGCUUAAAAGAACAUUCAAAUGGAAAUGUUCUGCUUGGCUCAACACAAACAACAAGUGCAGGC----AAACAAACUGCAUUUAUUUAGUUAACAAAAAAAAAAA ......-..(((((..((((....((((((((........))))))))....)))..............((((((((..----.......)))))))).....)..)))))......... ( -24.20) >DroPse_CAF1 45319 101 + 1 AGAAGA-AAUUGUUUUCGCUUAAAAGAACAUUCAAAUGGAAAUGUUCUGCUUGGCUCAACACAAACAACAA------------------GCUGCAUUUAUUUGGUUAGCAAACAAAAAAU ......-..((((((..(((....((((((((........))))))))....)))................------------------((((.(((.....)))))))))))))..... ( -17.30) >DroEre_CAF1 50384 120 + 1 AGAAAAGAAUUGUUUUCGCUUAAAAGAACAUUCAAAUGGAAAUGUUCUGCUUGGCUCAACACAAACAACCAGUGCAGGCGCACACACAAACUGCAUUUAUUUGGUUAGCAGGAAAAAUGG ............((((((((....((((((((........))))))))....)))...........(((((((((((.............)))))).....))))).....))))).... ( -24.82) >DroYak_CAF1 51375 119 + 1 AGAAAAGAAUUGUUUUCGCUUAAAAGAACAUUCAAAUGGAAAUGUUCUGCUUGGCUCAACACAAACAACAAGUGCAGGCGCACACACAAACUGCAUUUAUUUAGUUAGCAAGCAAAAUG- .........((((((.((((....((((((((........))))))))....)))..............((((((((.............)))))))).........).))))))....- ( -23.92) >DroAna_CAF1 39349 120 + 1 AGAAAGGAAUUGUUUUCGCUUAAAAGAACAUUCAAAUGGAAAUGUUCUGCUUGGCUCAACACAAACAACGAAUGCAGGCGCGCACACAAACUGCAUUUAUUUAGUUAGCAAACAAAAAGG .........((((((.((((....((((((((........))))))))....)))..............((((((((.............)))))))).........).))))))..... ( -24.32) >consensus AGAAAAGAAUUGUUUUCGCUUAAAAGAACAUUCAAAUGGAAAUGUUCUGCUUGGCUCAACACAAACAACAAGUGCAGGCGCACACACAAACUGCAUUUAUUUAGUUAGCAAACAAAAAAG .........(((((..((((....((((((((........))))))))....)))..............((((((((.............)))))))).....)..)))))......... (-20.43 = -21.10 + 0.67)


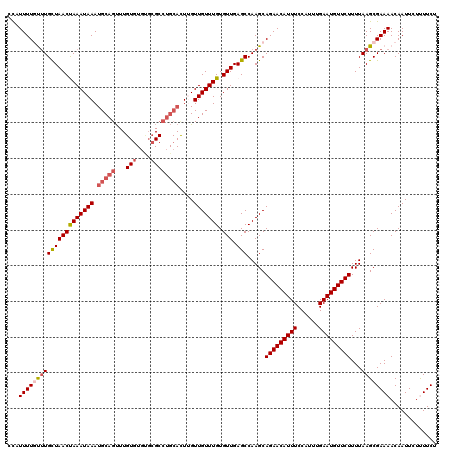
| Location | 18,406,612 – 18,406,732 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.19 |
| Mean single sequence MFE | -25.95 |
| Consensus MFE | -21.79 |
| Energy contribution | -22.83 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.599792 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18406612 120 - 23771897 CUAUUUUGUUUGCUAACUAAAUAAAUGCAGUUUGUGUGUGCGCCUGCACUUGUUGUUUGUGUUGAGCCAAGCAGAACAUUUCCAUUUGAAUGUUCUUUUAAGCGAAAACAAUUCUUUUCU ...(((((((((((((((((((((.(((((...(((....))))))))....))))))).))).))).....(((((((((......)))))))))...))))))))............. ( -27.10) >DroVir_CAF1 122606 115 - 1 UUUUUUUUUUUGUUAACUAAAUAAAUGCAGUUUGUUU----GCCUGCACUUGUUGUUUGUGUUGAGCCAAGCAGAACAUUUCCAUUUGAAUGUUCUUUUAAGCGAAAACAAUU-UCUUCU ......((((((((.....(((((.(((((.......----..))))).)))))(((((.(.....))))))(((((((((......)))))))))....)))))))).....-...... ( -22.90) >DroPse_CAF1 45319 101 - 1 AUUUUUUGUUUGCUAACCAAAUAAAUGCAGC------------------UUGUUGUUUGUGUUGAGCCAAGCAGAACAUUUCCAUUUGAAUGUUCUUUUAAGCGAAAACAAUU-UCUUCU ....((((((((.....)))))))).((.((------------------(((..((((.....)))))))))(((((((((......))))))))).....))((((....))-)).... ( -20.90) >DroEre_CAF1 50384 120 - 1 CCAUUUUUCCUGCUAACCAAAUAAAUGCAGUUUGUGUGUGCGCCUGCACUGGUUGUUUGUGUUGAGCCAAGCAGAACAUUUCCAUUUGAAUGUUCUUUUAAGCGAAAACAAUUCUUUUCU ....((((((((((((((((((((.(((((...(((....))))))))....))))))).))).))).....(((((((((......)))))))))....)).)))))............ ( -27.30) >DroYak_CAF1 51375 119 - 1 -CAUUUUGCUUGCUAACUAAAUAAAUGCAGUUUGUGUGUGCGCCUGCACUUGUUGUUUGUGUUGAGCCAAGCAGAACAUUUCCAUUUGAAUGUUCUUUUAAGCGAAAACAAUUCUUUUCU -..(((((((((((((((((((((.(((((...(((....))))))))....))))))).))).))).....(((((((((......)))))))))...))))))))............. ( -29.50) >DroAna_CAF1 39349 120 - 1 CCUUUUUGUUUGCUAACUAAAUAAAUGCAGUUUGUGUGCGCGCCUGCAUUCGUUGUUUGUGUUGAGCCAAGCAGAACAUUUCCAUUUGAAUGUUCUUUUAAGCGAAAACAAUUCCUUUCU ..((((((((((((((((((((((((((((...(((....)))))))))...))))))).))).))).....(((((((((......)))))))))...)))))))))............ ( -28.00) >consensus CCAUUUUGUUUGCUAACUAAAUAAAUGCAGUUUGUGUGUGCGCCUGCACUUGUUGUUUGUGUUGAGCCAAGCAGAACAUUUCCAUUUGAAUGUUCUUUUAAGCGAAAACAAUUCUUUUCU ...(((((((((((((((((((((.(((((...(((....))))))))....))))))).))).))).....(((((((((......)))))))))...))))))))............. (-21.79 = -22.83 + 1.04)

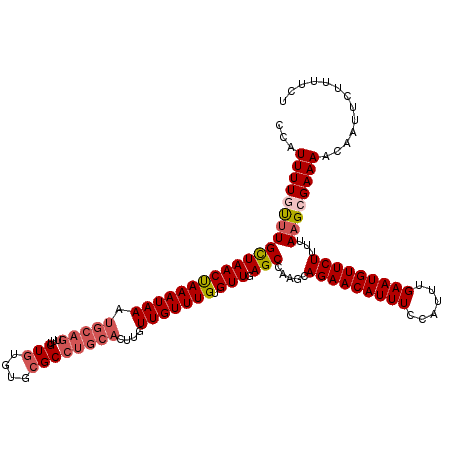
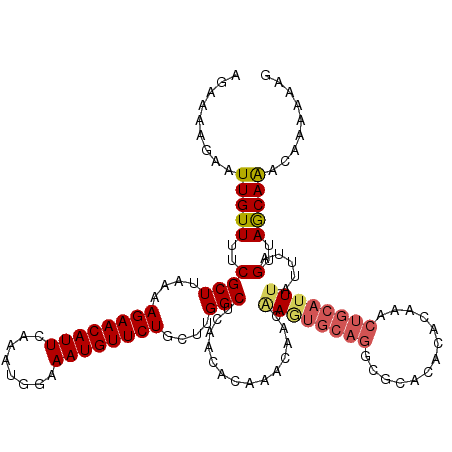
| Location | 18,406,652 – 18,406,756 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 86.78 |
| Mean single sequence MFE | -22.18 |
| Consensus MFE | -21.18 |
| Energy contribution | -21.15 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.894682 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18406652 104 - 23771897 -UUUGCUCCUUUUCACUU-------UUUUUUU---CUAUUUUGUUUGCUAACUAAAUAAAUGCAGUUUGUGUGUGCGCCUGCACUUGUUGUUUGUGUUGAGCCAAGCAGAACAUU -.................-------.......---...(((((((((((((((((((((.(((((...(((....))))))))....))))))).))).)).))))))))).... ( -21.40) >DroSec_CAF1 42897 110 - 1 -UUUGCUCCUUUGCUCCUU-UCCCGUUUUUUU---CUAUUUUGUUUGCUAACUAAAUAAAUGCAGUUUGUGUGUGCGCCUGCACUUGUUGUUUGUGUUGAGCCAAGCAGAACAUU -...((......)).....-............---...(((((((((((((((((((((.(((((...(((....))))))))....))))))).))).)).))))))))).... ( -22.60) >DroSim_CAF1 42051 111 - 1 -UUUGCUCCUUUGCUCCUUUUCCCUUUUUUUU---CUAUUUUGUUUGCUAACUAAAUAAAUGCAGUUUGUGUGUGCGCCUGCACUUGUUGUUUGUGUUGAGCCAAGCAGAACAUU -...((......))..................---...(((((((((((((((((((((.(((((...(((....))))))))....))))))).))).)).))))))))).... ( -22.60) >DroEre_CAF1 50424 103 - 1 -UUUGCUGCUUU--------UCCCUUUUUUCC---CCAUUUUUCCUGCUAACCAAAUAAAUGCAGUUUGUGUGUGCGCCUGCACUGGUUGUUUGUGUUGAGCCAAGCAGAACAUU -....((((((.--------............---...........(((((((((((((.(((((...(((....))))))))....))))))).))).))).))))))...... ( -20.91) >DroYak_CAF1 51415 101 - 1 -UUUGCUCCUUU--------UCCCUUUUUUC-----CAUUUUGCUUGCUAACUAAAUAAAUGCAGUUUGUGUGUGCGCCUGCACUUGUUGUUUGUGUUGAGCCAAGCAGAACAUU -...........--------...........-----..(((((((((((((((((((((.(((((...(((....))))))))....))))))).))).)).))))))))).... ( -23.80) >DroAna_CAF1 39389 109 - 1 UUUUGCUCCUUUUUA------UCCUUUUUUUGCUCCCUUUUUGUUUGCUAACUAAAUAAAUGCAGUUUGUGUGCGCGCCUGCAUUCGUUGUUUGUGUUGAGCCAAGCAGAACAUU ...............------.................(((((((((((((((((((((((((((...(((....)))))))))...))))))).))).)).))))))))).... ( -21.80) >consensus _UUUGCUCCUUU_C______UCCCUUUUUUUU___CCAUUUUGUUUGCUAACUAAAUAAAUGCAGUUUGUGUGUGCGCCUGCACUUGUUGUUUGUGUUGAGCCAAGCAGAACAUU ......................................(((((((((((((((((((((.(((((...(((....))))))))....))))))).))).)).))))))))).... (-21.18 = -21.15 + -0.03)

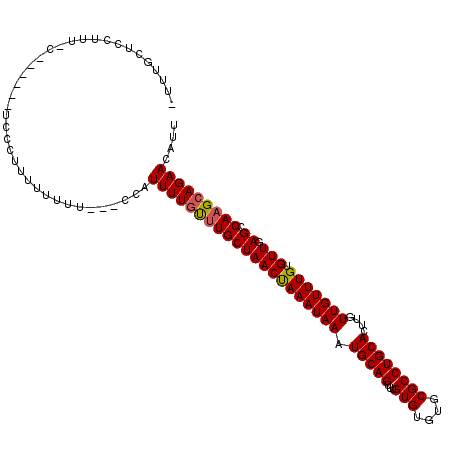
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:31:26 2006