
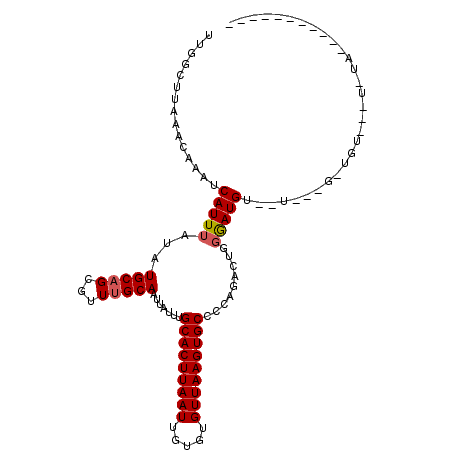
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 18,398,271 – 18,398,367 |
| Length | 96 |
| Max. P | 0.917403 |

| Location | 18,398,271 – 18,398,367 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 79.32 |
| Mean single sequence MFE | -24.49 |
| Consensus MFE | -13.67 |
| Energy contribution | -13.87 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.692557 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18398271 96 + 23771897 UUGGCUUAAACAAAUCAUUUAUAUGCAGCGCUCGCAAUUAUUUGCACUUAAUUGUGUGUUAAGUGCCCCAGACUGGGAUGU--UC--GGUGUGCAUAUAG---------- ...................((((((((.((((.(((.......(((((((((.....)))))))))(((.....))).)))--..--)))))))))))).---------- ( -29.00) >DroPse_CAF1 34899 82 + 1 UUGGCUUAAACAAAUCAUUUAUAUGCAGCGUUUGCAAUUAUUUGCACUUAAUUGUGUGUUAAGUGCCCCAGACUGGGAUGU--U-------------------------- ........((((...........(((((...))))).......(((((((((.....)))))))))(((.....))).)))--)-------------------------- ( -17.70) >DroGri_CAF1 178625 107 + 1 UCGGCUUAAACAAAUCAUUUAUAUGCAGCGUUUGCAAUUAUUUGCACUUAAUUGUGUGUUAAGUGCCC-AGCCAGCAAUGCCUUC--GCUGUUGUUGUAGAGCCUGACGC ..(((((.(((((..(((....)))(((((...(((.......(((((((((.....)))))))))..-.(....)..)))...)--)))))))))...)))))...... ( -29.50) >DroWil_CAF1 445057 95 + 1 UUAGCUUAAACAAAUCAUUUAUAUGCAGCGUUUGCAAUUAUUUGCACUUAAUUGUGUGUUAAGUGCCCAAGCCUGGGAUGG--AU--GAUGCGA-UGCGA---------- ...((........(((((((((.(.(((.(((((.........(((((((((.....))))))))).)))))))).)))))--))--)))....-.))..---------- ( -25.02) >DroAna_CAF1 31574 98 + 1 UUAGCUUAAACAAAUCAUUUAUAUGCAGCGCUUGCAAUUAUUUGCACUUAAUUGUGUGUUAAGUGCCCCAGACUGGGAUGC--AAGCAGUAUACACAUAC---------- .....................(((((...(((((((.......(((((((((.....)))))))))(((.....))).)))--)))).))))).......---------- ( -28.00) >DroPer_CAF1 34732 82 + 1 UUGGCUUAAACAAAUCAUUUAUAUGCAGCGUUUGCAAUUAUUUGCACUUAAUUGUGUGUUAAGUGCCCCAGACUGGGAUGU--U-------------------------- ........((((...........(((((...))))).......(((((((((.....)))))))))(((.....))).)))--)-------------------------- ( -17.70) >consensus UUGGCUUAAACAAAUCAUUUAUAUGCAGCGUUUGCAAUUAUUUGCACUUAAUUGUGUGUUAAGUGCCCCAGACUGGGAUGU__U___G_UGU___U_UA___________ ...............(((((...(((((...))))).......(((((((((.....))))))))).........))))).............................. (-13.67 = -13.87 + 0.19)

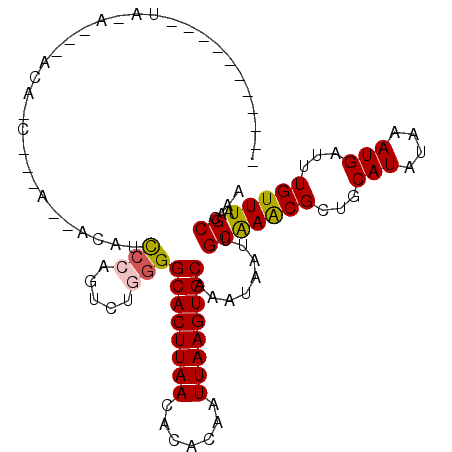
| Location | 18,398,271 – 18,398,367 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 79.32 |
| Mean single sequence MFE | -21.98 |
| Consensus MFE | -13.60 |
| Energy contribution | -13.60 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.917403 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18398271 96 - 23771897 ----------CUAUAUGCACACC--GA--ACAUCCCAGUCUGGGGCACUUAACACACAAUUAAGUGCAAAUAAUUGCGAGCGCUGCAUAUAAAUGAUUUGUUUAAGCCAA ----------.(((((((((.((--(.--....(((.....)))((((((((.......)))))))).........)).).).))))))))................... ( -23.20) >DroPse_CAF1 34899 82 - 1 --------------------------A--ACAUCCCAGUCUGGGGCACUUAACACACAAUUAAGUGCAAAUAAUUGCAAACGCUGCAUAUAAAUGAUUUGUUUAAGCCAA --------------------------.--....(((.....)))((((((((.......)))))))).......((((.....))))..(((((.....)))))...... ( -16.00) >DroGri_CAF1 178625 107 - 1 GCGUCAGGCUCUACAACAACAGC--GAAGGCAUUGCUGGCU-GGGCACUUAACACACAAUUAAGUGCAAAUAAUUGCAAACGCUGCAUAUAAAUGAUUUGUUUAAGCCGA ......((((..((((((.((((--((.....))))))..)-).((((((((.......)))))))).......((((.....))))..........))))...)))).. ( -27.20) >DroWil_CAF1 445057 95 - 1 ----------UCGCA-UCGCAUC--AU--CCAUCCCAGGCUUGGGCACUUAACACACAAUUAAGUGCAAAUAAUUGCAAACGCUGCAUAUAAAUGAUUUGUUUAAGCUAA ----------..((.-..(((..--..--(((.(....)..)))((((((((.......)))))))).......)))(((((...(((....)))...)))))..))... ( -18.60) >DroAna_CAF1 31574 98 - 1 ----------GUAUGUGUAUACUGCUU--GCAUCCCAGUCUGGGGCACUUAACACACAAUUAAGUGCAAAUAAUUGCAAGCGCUGCAUAUAAAUGAUUUGUUUAAGCUAA ----------.((((((((...(((((--(((.(((.....)))((((((((.......)))))))).......)))))))).))))))))................... ( -30.90) >DroPer_CAF1 34732 82 - 1 --------------------------A--ACAUCCCAGUCUGGGGCACUUAACACACAAUUAAGUGCAAAUAAUUGCAAACGCUGCAUAUAAAUGAUUUGUUUAAGCCAA --------------------------.--....(((.....)))((((((((.......)))))))).......((((.....))))..(((((.....)))))...... ( -16.00) >consensus ___________UA_A___ACA_C___A__ACAUCCCAGUCUGGGGCACUUAACACACAAUUAAGUGCAAAUAAUUGCAAACGCUGCAUAUAAAUGAUUUGUUUAAGCCAA .................................(((.....)))((((((((.......))))))))........(((((((...(((....)))...)))))..))... (-13.60 = -13.60 + 0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:31:20 2006