
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 18,363,924 – 18,364,092 |
| Length | 168 |
| Max. P | 0.903971 |

| Location | 18,363,924 – 18,364,020 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 74.34 |
| Mean single sequence MFE | -18.94 |
| Consensus MFE | -8.82 |
| Energy contribution | -9.49 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.47 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.584157 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
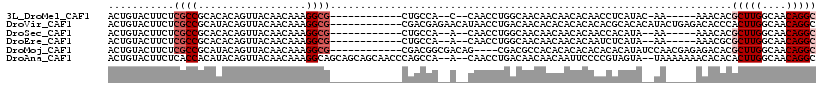
>3L_DroMel_CAF1 18363924 96 - 23771897 ACUGUACUUCUCGCCGCACACAGUUACAACAAAGGCG------------CUGCCA--C--CAACCUGGCAACAACAACACAACCUCAUAC-AA-----AAACACGCUUGGCAACAGGC ..((((.....((((..................))))------------.(((((--.--.....))))).................)))-).-----......(((((....))))) ( -20.27) >DroVir_CAF1 84260 106 - 1 ACUGUACUUCUCGCCGCAUACAGUUACAACAAAGGCG------------CGACGAGAACAUAACCUGACAACACACACACACACGCACACAUACUGAGACACCCACUUGGCAACAGGC .((((..((((((((((.....((....))....)))------------.).))))))...................................(..((.......))..)..)))).. ( -16.10) >DroSec_CAF1 632 95 - 1 ACUGUACUUCUCGCCGCACACAGUUACAACAAAGGCG------------CUGCCA--A--CAACCUGGCAACAACAACACAACCACAUA--AA-----AAACACGCUUGGCAACAGGC ..(((......((((..................))))------------.(((((--.--.....))))))))................--..-----......(((((....))))) ( -19.47) >DroEre_CAF1 8194 95 - 1 ACUGUACUUCUCGCCGCACACAGUUACAACAAAGGCG------------CUGCCA--A--CAACCUGGCAACAACAACACAAUCUCAUA--AA-----AAACGCGCUUGGCAACAGGC ..(((......((((..................))))------------.(((((--.--.....))))))))................--..-----......(((((....))))) ( -19.47) >DroMoj_CAF1 116030 102 - 1 ACUGUACUUCUCGCCGCAUACAGUUACAACAAAGGCG------------CGACGGCGACAG----CGACGCCACACACACACACACAUAUCCAACGAGAGACACGCUUGGCAACAGGC ...((..((((((.........((....))...((((------------((..(....)..----)).))))......................))))))..))(((((....))))) ( -24.90) >DroAna_CAF1 693 112 - 1 ACUGUACUUCUCACCACAUACAGUUACAACAAAGGCAGCAGCAGCAACCCAGCCA--A--CAACCUGACAACAACAAUUCCCCGUAGUA--UAAAAAAACACACACUUGGCAACAGGC ((((((............))))))(((.((...((..((....))..))(((...--.--....)))................)).)))--..............((((....)))). ( -13.40) >consensus ACUGUACUUCUCGCCGCACACAGUUACAACAAAGGCG____________CUGCCA__A__CAACCUGACAACAACAACACAACCUCAUA__AA_____AAACACGCUUGGCAACAGGC ...........((((..................))))...................................................................(((((....))))) ( -8.82 = -9.49 + 0.67)

| Location | 18,363,956 – 18,364,060 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.91 |
| Mean single sequence MFE | -34.00 |
| Consensus MFE | -20.72 |
| Energy contribution | -20.06 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.605861 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
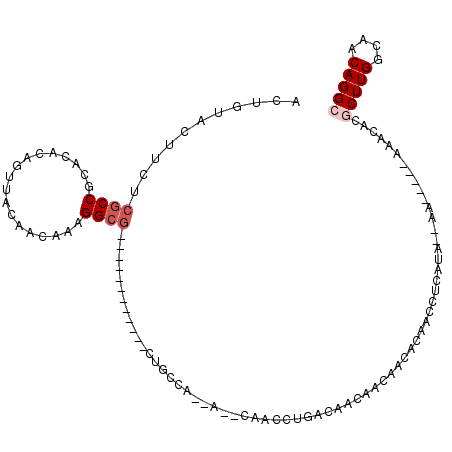
>3L_DroMel_CAF1 18363956 104 + 23771897 GUGUUGUUGUUGCCAGGUUG--G--UGGCAG------------CGCCUUUGUUGUAACUGUGUGCGGCGAGAAGUACAGUUAGCAUAUUAAUUGUAUUAAAUCGUCGGGGCAUGACAAUC .(((((((((..((.....)--)--..))))------------)(((((....(((......)))(((((..(((((((((((....)))))))))))...))))))))))..))))... ( -36.50) >DroVir_CAF1 84298 108 + 1 GUGUGUGUGUUGUCAGGUUAUGUUCUCGUCG------------CGCCUUUGUUGUAACUGUAUGCGGCGAGAAGUACAGUUAGCAUAUUAAUUGUAUUAAAUCGUCGUUGCAUGACAAAC .........((((((.((.(((.((((((((------------((.....((....))....))))))))))(((((((((((....))))))))))).......))).)).)))))).. ( -31.20) >DroSec_CAF1 663 104 + 1 GUGUUGUUGUUGCCAGGUUG--U--UGGCAG------------CGCCUUUGUUGUAACUGUGUGCGGCGAGAAGUACAGUUAGCAUAUUAAUUGUAUUAAAUCGUCGGUGCAUGACAAUC ..(((((..(((((((....--)--)))))(------------((((......(((......)))(((((..(((((((((((....)))))))))))...)))))))))))..))))). ( -35.50) >DroSim_CAF1 662 104 + 1 GUGUUGUUGUUGCCAGGUUG--U--UGGCAG------------CGCCUUUGUUGUAACUGUGUGUGGCGAGAAGUACAGUUAGCAUAUUAAUUGUAUUAAAUCGUCGGUGCAUGACAAUC ..(((((..(((((((....--)--)))))(------------((((.........((.....))(((((..(((((((((((....)))))))))))...)))))))))))..))))). ( -34.20) >DroMoj_CAF1 116068 104 + 1 GUGUGUGUGGCGUCG----CUGUCGCCGUCG------------CGCCUUUGUUGUAACUGUAUGCGGCGAGAAGUACAGUUAGCAUAUUAAUUGUAUUAAAUCGUCGUCGCAUGACAAAC ..((((((((((...----....))))).))------------)))............(((((((((((((((((((((((((....)))))))))))...)).))))))))).)))... ( -36.80) >DroAna_CAF1 729 116 + 1 GAAUUGUUGUUGUCAGGUUG--U--UGGCUGGGUUGCUGCUGCUGCCUUUGUUGUAACUGUAUGUGGUGAGAAGUACAGUUAGCAUAUUAAUUGUAUUUAAUCGUCAGGGCAUGACAAAC ...((((..(.(((.(((((--.--..((.((((.((....)).))))..))..))))).....((..((.((((((((((((....))))))))))))..))..)).))))..)))).. ( -29.80) >consensus GUGUUGUUGUUGCCAGGUUG__U__UGGCAG____________CGCCUUUGUUGUAACUGUAUGCGGCGAGAAGUACAGUUAGCAUAUUAAUUGUAUUAAAUCGUCGGUGCAUGACAAAC .....(((((((((((..........(((...............)))..........))).)).((((((..(((((((((((....)))))))))))...))))))......)))))). (-20.72 = -20.06 + -0.66)


| Location | 18,363,983 – 18,364,092 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.68 |
| Mean single sequence MFE | -32.79 |
| Consensus MFE | -20.31 |
| Energy contribution | -19.48 |
| Covariance contribution | -0.83 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.903971 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
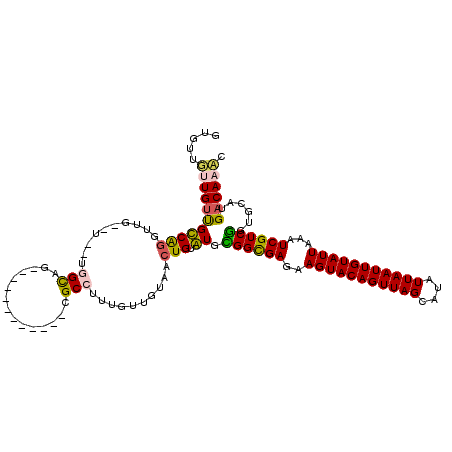
>3L_DroMel_CAF1 18363983 109 + 23771897 ---CGCCUUUGUUGUAACUGUGUGCGGCGAGAAGUACAGUUAGCAUAUUAAUUGUAUUAAAUCGUCGGGGCAUGACAAUCGUUUCUCAUCAAGG--------AGUUCGACAGAGAAAGAC ---...((((((((.((((.(.((.(((((..(((((((((((....)))))))))))...)))))((((.(((.....))).))))..)).).--------))))))))))))...... ( -32.70) >DroVir_CAF1 84329 112 + 1 ---CGCCUUUGUUGUAACUGUAUGCGGCGAGAAGUACAGUUAGCAUAUUAAUUGUAUUAAAUCGUCGUUGCAUGACAAACAUUU-UCAGCACAACGUCUGGAAGGCAAAAAGAGAG---- ---.((((((.((((...((((.(((((((..(((((((((((....)))))))))))...)))))))))))..))))......-.((((.....).)))))))))..........---- ( -33.60) >DroSim_CAF1 689 109 + 1 ---CGCCUUUGUUGUAACUGUGUGUGGCGAGAAGUACAGUUAGCAUAUUAAUUGUAUUAAAUCGUCGGUGCAUGACAAUCGUUUUUCACCAAGG--------AGUUCGACAGAGAAAGAC ---...((((((((.((((.(....(((((..(((((((((((....)))))))))))...)))))((((...(((....)))...))))..).--------))))))))))))...... ( -34.30) >DroYak_CAF1 6605 108 + 1 ---CGCCUUUGUUGUAACUGUGUGCGGCGAGAAGUACAGUUAGCAUAUUAAUUGUAUUAAAUCGUCGGAGCAUGACACUCGUUU-UCACCAAGG--------AGUUCGACGGAGAAAGAC ---...((((((((.((((.(.((((((((..(((((((((((....)))))))))))...))))))(((.(((.....))).)-))..)).).--------))))))))))))...... ( -31.60) >DroMoj_CAF1 116095 112 + 1 ---CGCCUUUGUUGUAACUGUAUGCGGCGAGAAGUACAGUUAGCAUAUUAAUUGUAUUAAAUCGUCGUCGCAUGACAAACAUUU-UCAACGCAACGUCUAGAAGGCAAAACAGGAG---- ---.(((((((((((...(((((((((((((((((((((((((....)))))))))))...)).))))))))).))).......-.....))))).....))))))..........---- ( -31.86) >DroAna_CAF1 765 111 + 1 UGCUGCCUUUGUUGUAACUGUAUGUGGUGAGAAGUACAGUUAGCAUAUUAAUUGUAUUUAAUCGUCAGGGCAUGACAAACGGUUUCAGCCCAAG--------UGUUGGACAGAGAAGG-C ....((((((((((.((((((....(((...((((((((((((....)))))))))))).)))((((.....))))..)))))).))))(((..--------...))).....)))))-) ( -32.70) >consensus ___CGCCUUUGUUGUAACUGUAUGCGGCGAGAAGUACAGUUAGCAUAUUAAUUGUAUUAAAUCGUCGGGGCAUGACAAACGUUU_UCACCAAAG________AGUUCGACAGAGAAAG_C ......(((((((.....(((...((((((..(((((((((((....)))))))))))...))))))..)))((.....))..........................)))))))...... (-20.31 = -19.48 + -0.83)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:31:13 2006