| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 18,361,322 – 18,361,427 |
| Length | 105 |
| Max. P | 0.574807 |

| Location | 18,361,322 – 18,361,427 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.33 |
| Mean single sequence MFE | -20.32 |
| Consensus MFE | -17.58 |
| Energy contribution | -17.31 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.574807 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
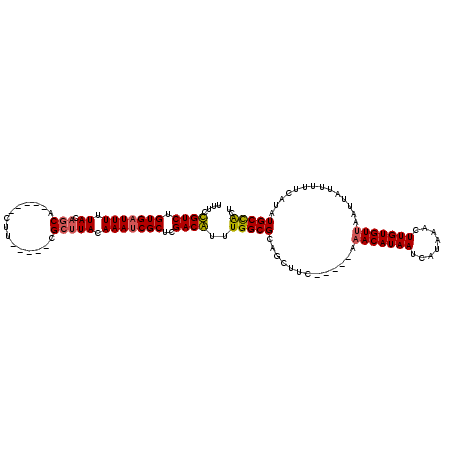
>3L_DroMel_CAF1 18361322 105 + 23771897 UUUUCGUCUGUGAUUUUUACAGCA-----CUU-----CGCUUACAAAUCGCUCGACGUUUGGCGCAGCUUC-----AAACAUAAUCAUAAACUUGUGUUAAUUAUUUUUCAUAUGCCACU ....((((.(((((((.((.(((.-----...-----.))))).)))))))..))))..(((((.......-----.(((((((........)))))))...(((.....)))))))).. ( -20.30) >DroPse_CAF1 171445 115 + 1 GUUCUGUCUGUGAUUUUUACAGCU-----CCUUCGGCUGCUUACAAAUCGCUCGACAUUUGGCGCAACUUCCACUCGAACAUAAUCAUAAACUUGUGUUAAUUAUUUUUCAUAUGCUGCU ....((((.(((((((.((.(((.-----.(....)..))))).)))))))..))))...((((((...........(((((((........)))))))..............))).))) ( -22.71) >DroSec_CAF1 145351 105 + 1 UUUCCGUCUGUGAUUUUUAGAGCA-----CUU-----CGCUUACAAAUCGCUCGACGUUUGGCGCAGCUUC-----AAACAUAAUCAUAAACUUGUGUUAAUUAUUUUUCAUAUGCCACU ....((((.(((((((...((((.-----...-----.))))..)))))))..))))..(((((.......-----.(((((((........)))))))...(((.....)))))))).. ( -21.40) >DroSim_CAF1 156354 105 + 1 UUUCCGUCUGUGAUUUUUACAGCA-----CUU-----CGCUUACAAAUCGCUCGACGUUUGGCGCAGCUUC-----AAACAUAAUCAUAAACUUGUGUUAAUUAUUUUUCAUAUGCCACU ....((((.(((((((.((.(((.-----...-----.))))).)))))))..))))..(((((.......-----.(((((((........)))))))...(((.....)))))))).. ( -20.50) >DroAna_CAF1 141888 109 + 1 UUUCUGUCUGUGCUUUUUAUUACAACUGCUUU-----CGCUUACAAAUCGCUCGACAUUUGGCGCAGCUUC-----A-ACAUAAUCAUAAACUUGUGUUAAUUAUUUUUCAUAUGCCAAU .....(.((((((..............((...-----.))...(((((........))))))))))))...-----(-((((((........)))))))..................... ( -14.30) >DroPer_CAF1 172436 115 + 1 GUUCUGUCUGUGAUUUUUACAGCU-----CCUUCGGCUGCUUACAAAUCGCUCGACAUUUGGCGCAACUUCCACUCGAACAUAAUCAUAAACUUGUGUUAAUUAUUUUUCAUAUGCUGCU ....((((.(((((((.((.(((.-----.(....)..))))).)))))))..))))...((((((...........(((((((........)))))))..............))).))) ( -22.71) >consensus UUUCCGUCUGUGAUUUUUACAGCA_____CUU_____CGCUUACAAAUCGCUCGACAUUUGGCGCAGCUUC_____AAACAUAAUCAUAAACUUGUGUUAAUUAUUUUUCAUAUGCCACU ....((((.(((((((.((.(((...............))))).)))))))..))))..(((((.............(((((((........)))))))..............))))).. (-17.58 = -17.31 + -0.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:31:10 2006