
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 18,360,886 – 18,360,981 |
| Length | 95 |
| Max. P | 0.998853 |

| Location | 18,360,886 – 18,360,981 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 76.14 |
| Mean single sequence MFE | -24.05 |
| Consensus MFE | -14.95 |
| Energy contribution | -15.95 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.62 |
| SVM decision value | 2.02 |
| SVM RNA-class probability | 0.985854 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
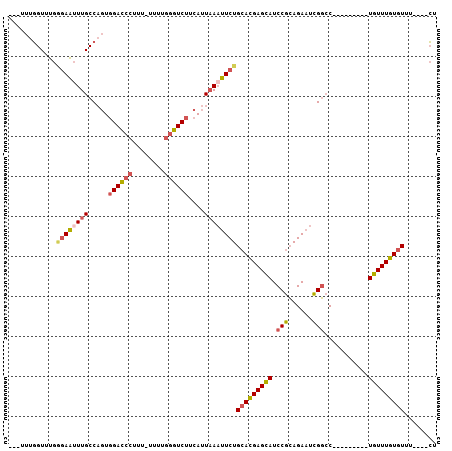
>3L_DroMel_CAF1 18360886 95 + 23771897 ---UUUGGUUUGGGAAUUUGCCAGUGGACCCUUUUUUUUGGGUCUUCAUUAAUUUCUGCACGAGUAUCCGCAGAAUCGGAC---------UAUUUGUGUUCCUUCCU ---...((...((((........(.((((((........)))))).)..........(((((((((((((......)))).---------))))))))))))).)). ( -29.80) >DroSec_CAF1 144929 90 + 1 ---UUUGGUUUGGGAAUUUGCCAGUGGACCCUUU-UUUUGGGUCUUCAUUAAAUUCUGCACGAGCAUCCGCAGAAUUGGCC---------UGUUUGUGUUU----CU ---...((..((.((((..(((.(.((((((...-....)))))).)....((((((((..........))))))))))).---------.)))).))..)----). ( -27.00) >DroSim_CAF1 155932 90 + 1 ---UUUGGUUUGAGAAUUUGCCAGUGGACCCUUU-UUUUGGGUCUUCAUUAAAUUCUGCACGAGCAUCCGCAGAAUUGGCC---------UGUUUGUGUUU----CU ---.........((((((((.....((((((...-....))))))....))))))))(((((((((.(((......)))..---------)))))))))..----.. ( -26.70) >DroAna_CAF1 141446 97 + 1 UUCUUUUGUCUG-CAGAUUGCCUUUCGAUCCUUC-UUU---GUCUUCAUUUAAUUCUGAAGGAGCAUCCGGAGA-UCGUCCCCACUUUAUUGUUUUUAUUU----GU ...(((((..((-(.(((((.....)))))....-...---.((((((........)))))).)))..))))).-..........................----.. ( -12.70) >consensus ___UUUGGUUUGGGAAUUUGCCAGUGGACCCUUU_UUUUGGGUCUUCAUUAAAUUCUGCACGAGCAUCCGCAGAAUCGGCC_________UGUUUGUGUUU____CU ............((((((((.....((((((........))))))....))))))))(((((((((.(((......)))...........)))))))))........ (-14.95 = -15.95 + 1.00)


| Location | 18,360,886 – 18,360,981 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 76.14 |
| Mean single sequence MFE | -21.23 |
| Consensus MFE | -12.85 |
| Energy contribution | -14.85 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.87 |
| Structure conservation index | 0.61 |
| SVM decision value | 3.25 |
| SVM RNA-class probability | 0.998853 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
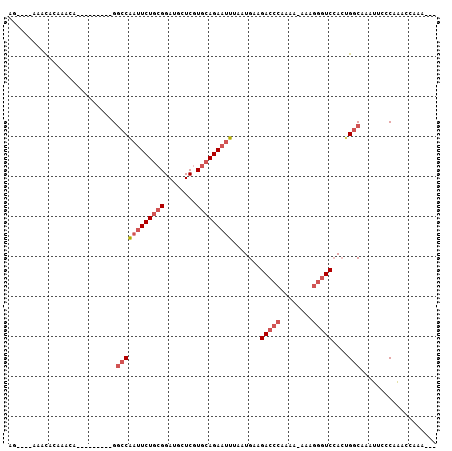
>3L_DroMel_CAF1 18360886 95 - 23771897 AGGAAGGAACACAAAUA---------GUCCGAUUCUGCGGAUACUCGUGCAGAAAUUAAUGAAGACCCAAAAAAAAGGGUCCACUGGCAAAUUCCCAAACCAAA--- .((..((((......((---------((....((((((((.....).))))))).........(((((........))))).)))).....))))....))...--- ( -21.80) >DroSec_CAF1 144929 90 - 1 AG----AAACACAAACA---------GGCCAAUUCUGCGGAUGCUCGUGCAGAAUUUAAUGAAGACCCAAAA-AAAGGGUCCACUGGCAAAUUCCCAAACCAAA--- ..----...........---------.(((((((((((((.....).))))))))).......(((((....-...)))))....)))................--- ( -22.30) >DroSim_CAF1 155932 90 - 1 AG----AAACACAAACA---------GGCCAAUUCUGCGGAUGCUCGUGCAGAAUUUAAUGAAGACCCAAAA-AAAGGGUCCACUGGCAAAUUCUCAAACCAAA--- ..----...........---------.(((((((((((((.....).))))))))).......(((((....-...)))))....)))................--- ( -22.30) >DroAna_CAF1 141446 97 - 1 AC----AAAUAAAAACAAUAAAGUGGGGACGA-UCUCCGGAUGCUCCUUCAGAAUUAAAUGAAGAC---AAA-GAAGGAUCGAAAGGCAAUCUG-CAGACAAAAGAA ..----..................((((....-.))))(..(((..(((((........)))))..---..(-((..(.((....)))..))))-))..)....... ( -18.50) >consensus AG____AAACACAAACA_________GGCCAAUUCUGCGGAUGCUCGUGCAGAAUUUAAUGAAGACCCAAAA_AAAGGGUCCACUGGCAAAUUCCCAAACCAAA___ ...........................((((((((((((........))))))))).......(((((........)))))....)))................... (-12.85 = -14.85 + 2.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:31:09 2006